Creative Biolabs provides the unique ecotin library construction service through our innovative Hi-Affi™ phage display platform. This platform offers the ability to construct high-affinity scaffold libraries which can obtain 100% precise mutant and over 1010 diversity.
Ecotin is a group of dimeric serine protease inhibitors found in the periplasm of Escherichia coli and related Gram-negative bacteria that have been shown to be potent inhibitors of many trypsin-fold serine proteases of widely varying substrate specificity. Serine proteases are a large family involved in many important biological processes, such as blood coagulation, fibrinolysis, complement pathways, viral maturation, apoptosis, and cancer. In this way, the investigation of natural serine protease inhibitors is an important way to understand the mechanism of serine proteases and develop the inhibitors to become useful therapeutics, especially for the cancer therapy.
As a potent inhibitor of a surprisingly wide range of serine proteases, ecotin is a β-sandwich scaffold designed for serine protease inhibitor selection. All types of ecotins have shown the ability to inhibit pancreatic digestive peptidases trypsin and chymotrypsin, while they can also selectively inhibit several proteases, such as blood peptidases factor Xa, elastase, thrombin and human urokinase-type plasminogen activator (uPA). For instance, uPA is a serine protease that plays an active role in cancer metastasis and tumor invasion, it is considered that the library of phage-displayed ecotin variants could be a useful tool for isolating a high-affinity inhibitor of uPA, which may contribute to the development of anti-tumor drug and cancer inhibitor. In this way, ecotin is possible to play a key role in the treatment of serine protease-related diseases.
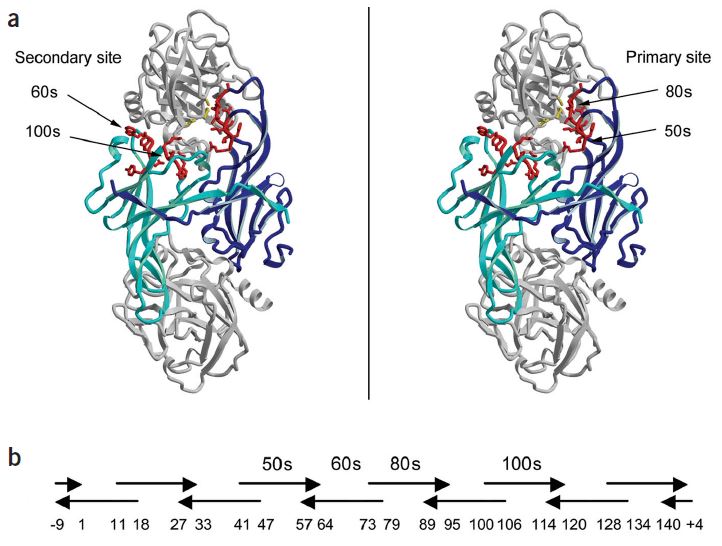
Fig.1 Positioning of ecotin surface loops in crystal structure and during synthetic shuffling. (Stoop and Craik, 2003)
In order to introduce the specificity in the ecotin scaffold, Creative Biolabs can generate custom ecotin libraries based on the mutagenesis for the four surface loops which are the 50s (position 51-54), 60s (positions 67-71), 80s (positions 81-86), and 100s (positions 108-112). Maximum 2020 (~1026) possibilities enable sufficient flexibility and diversity during the design of the novel ecotin library.
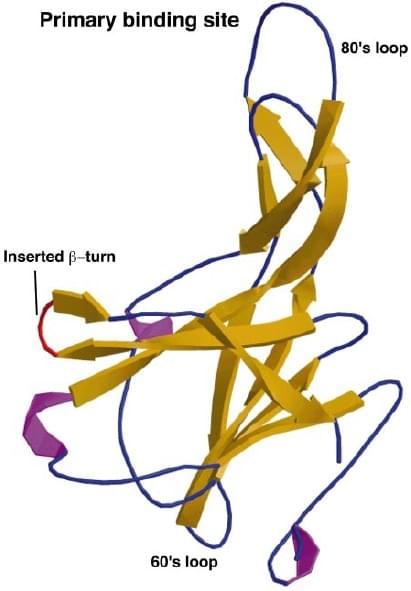
Fig. 2 Ribbon plot of a 2.0 Å structure of mEcotin, showing β-sheets in yellow, helices in magenta, and flexible loops and turns in blue. The introduced β-turn is labeled red. (Eggers et al. 2001)
Hi-Affi™ platform is developed by Creative Biolabs to combine common phage display technology with the trimer codon technology and NNK method. Compared with traditional phage display technology, which is an exogenous gene expression method through fusing the target genes to bacteriophage coat proteins then displaying on the phage surfaces to select specific binders, Hi-Affi™ phage display technology shows improved library affinity and diversity. The platform is more suitable for sorting and isolating the high-affinity protein or peptide targets.
With extensive experience in the field of scaffold library construction, Creative Biolabs has generated over 50 different kinds of scaffold libraries for our customers all over the world. Our scientists are committed to offering cost-effective service with the highest quality to assist our clients’ research project constantly moving forward. If you are interested in our ecotin library construction service, please do not hesitate to inquire us for more details.
References
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.