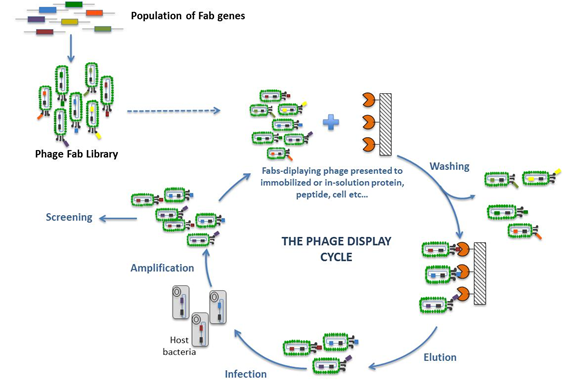
The phage display technology, first described over a decade ago, has opened new windows in the production of recombinant antibody fragments with the promise of trapping the naive immune repertoire in vitro. Phage display is a term describing display of foreign (poly) peptides on the surface of phage particle. The technique differs from conventional expression systems, however, in that the foreign gene sequence is spliced into the gene for one of the phage coat proteins, so that the foreign amino acid sequence is genetically fused to the endogenous ones of the coat protein to make a hybrid “fusion” protein. Using recombinant DNA technology, collections of billions of certain ligands (e.g., peptides or proteins, antibody fragments), cDNA-encoded or gene fragment-proteins presented on phage (so called phage display libraries) can be constructed and surveyed for specific affinity or activity (Fig. 1).
Fig. 1. Phage display cycle. DNA encoding for millions of variants of certain ligands (e.g., fab,peptides, proteins, or fragments thereof) is batch-cloned into the phage genome as part of one of the phage coat proteins (pIII, pVI, or pVIII). Large libraries containing millions of different ligands can be obtained by force-cloning in E. coli. From these repertoires, phage carrying specific-binding ligands can be isolated by a series of recursive cycles of selection on Ag(agar plate), each of which involves binding, washing, elution, and amplification.
Antibody was the first protein to be displayed successfully on the surface of phage. For therapeutic monoclonal antibodies, phage display system is typically used for three applications: identification and isolation of target specific antibodies either from non-immune, naïve, libraries or from libraries derived from genetic engineering of antibodies or immunized animals to optimize them for increased higher affinity or improved biophysical characteristics.
The main features of phage display technology in antibody production includes: sequences cloning of antibody library; selection of phage vector that can make large antibody libraries; linkage of the gene sequence with the displayed antibody; a screening (or selection) of identifying antibody variants that uniquely bind the target of choice; amplification of the nucleic acid sequence; either without or with introduction of diversity; and production of soluble antibody for functional characterization. The widespread use of antibody phage display comes from its relative simplicity, durability, stability of the phage particles and the ability to generate large libraries suitable for both de novo discovery and antibody optimization.
The phage display process for discovering antibodies or proteins that bind specifically to a given target will be detailed in the following so will only be highlighted here. Selection of phage Antibody involves the sequential enrichment of specific binding phage from an excess of nonbinding clones, which is achieved by multiple rounds of phage binding to the target, washing to remove unbound phage, and elution to retrieve specific binding phage. The target protein is tagged with biotin; target ones can be bound with very high affinity to streptavidin-coated plates or beads. Typically the target protein is either immobilized on a surface or beads that can be washed to remove unbound phage.
Effective display formats for Antibodies(Abs) are scFv, Fabs, immunoglobulin variable fragments (Fvs) with an engineered intermolecular disulphide bond to stabilize the VH–VL pair and diabody fragments. Antibody libraries are constructed by reverse-transcribing and PCR amplifying genes encoding antigen-binding domains of heavy (VH) and light (VL) chains from lymphocyte total RNA. By combining different VH and VL, a process termed chain shuffling, unique antibody fragments are formed. The smaller size of the scFv format makes their libraries genetically more stable than Fab libraries. To display Fabs on phage, either the light or heavy chain is fused via its C-terminus to vector, the partner chain is expressed and secreted into the periplasmic space where chain association forms an intact Fabs. Over the past decade, several fully human antibody phage display libraries have been constructed using either Fab or scFv formats. There are two general formats used for phage display of antibodies based on where the antibody (ab) genes are inserted. In one, the display protein-ab fusion is encoded on a separate plasmid, a phagemid that is subsequently encapsulated into phage particles upon infection with helper phage. In this case, the number of pIII-fusion proteins in the mature phage is either zero or one (i.e. monovalent display), allowing high affinity binders to be more easily isolated due to the absence of avidity effects. In the second format, the ab and display protein gene sequences are fused and incorporated directly into the phage genome so that five copies of a pIII-antibody fusion protein are produced. This format works reasonably well for libraries and general screenings but falls short when seeking high affinity binders as these are difficult to distinguish from lower affinity binders that have apparent higher affinity resulting from the avidity effect created by the binding of five copies to the target.
To date, there are at least 25 therapeutic antibodies that are derived from phage display libraries approved for clinical use or in various stages of clinical trials. One advantage of phage display is that fully human antibody can be isolated. Furthermore, phage display offers greater control over the selection process than using animal immunization. Finally, compared to other in vitro selection methods such as cell display, phage display is more readily accessible as it does not require expensive equipment like fluorescence-activated cell sorter (FACS). One shortcoming of phage display is that selection of antibody fragments on phage is based on expression. Some antibodies can be toxic to E. coli and accumulate stop codons throughout their coding sequence during the phage display process and be eliminated. Because of phage volume and size, as well as the maximum efficiency of E. coli transformation, phage display libraries have been limited to about 1011 unique members. Hybridoma-based antibodies, on the other hand, are subject to in vivo “editing” which tends to reduce problematic antibody properties such as aggregation. One difference from hybridoma technology is that the output of most display systems include antibody fragments such as scFvs or Fabs that must be further manipulated genetically to yield complete IgGs which are often required for initial functional evaluation.
Creative Biolabs provides comprehensive antibody Fab/scFv production service by phage display to meet your biopharmaceutical and biotechnology goal. We have long-term devoted to the development and application of phage display technology.