Rapid and simple methods for the epitope characterization as well as the antibody cross-reactivity with other proteins are great needed to generate well-validated, protein-specific diagnostic, and therapeutic antibodies. Creative Biolabs has established state-of-the-art bacterial display platform for epitope mapping services.
Bacterial display (or bacteria display or bacterial surface display) is a protein engineering technique which displays libraries containing billions of diverse molecules polypeptides on the surface of bacteria and, subsequently, to identify rarely desired polypeptides using selection or high-throughput screening methods. The bacterial display is often coupled with magnetic-activated cell sorting (MACS) or fluorescence-activated cell sorting (FACS) techniques which allow real-time analysis of displayed peptide properties, including target-binding affinity, specificity, and peptide stability to proteases during screening.
Epitope Mapping by Bacterial Display
Several methods are frequently used for mapping epitopes, involving scanning with chemically synthesized peptides, expressing peptides on the surface of microorganisms (such as phage display, yeast display), X-ray crystallography, NMR, and alanine scanning. However, such efforts are labor-intense, with relatively low success rates.
Creative Biolabs provides bacterial display strategy for epitope mapping, and it has been demonstrated successful by using several scaffolds. This strategy provides clear advantages to previously described mapping efforts (such as using phage-displayed peptide libraries), which are not suitable for fluorescence-activated cell sorting (FACS). The bacterial display uses cells as hosts for the libraries, and it combines FACS and subsequent DNA pyrosequencing, which provide a highly streamlined approach for epitope mapping of both monoclonal and polyclonal antibodies. Besides, this method will likely provide a means to enhance the specificity of ligand panels, since specificity can be measured directly using FACS.
Epitope Mapping Approach
The bacterial display based epitope mapping approach constructs libraries of overlapping peptides. The library comprises totally randomized fragmentation of the gene encoding the corresponding antigen in combination with a homology search using the selected mimotopes. Gram-positive bacterium Staphylococcus carnosus and Gram-negative bacterium Escherichia coli can be used as hosts for display.
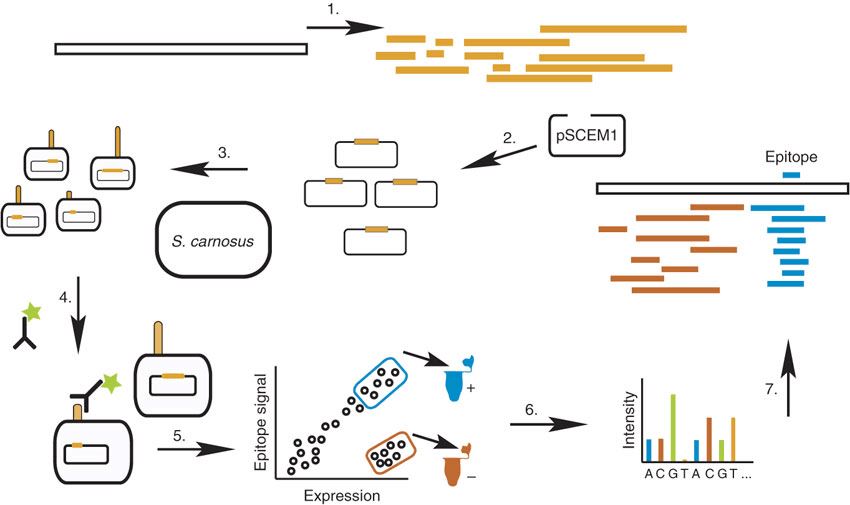 Figure 1. Schematic outline of the epitope mapping approach. (Rockberg et al. 2008)
Figure 1. Schematic outline of the epitope mapping approach. (Rockberg et al. 2008)
Brief Protocol of Bacterial Display based Epitope Mapping
For more detailed information, please feel free to contact us or directly sent us an inquiry.
References
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.