Creative Biolabs provides the PHD finger library construction service by rapid and high-throughput library generation platform Hi-Affi™. Built on the top of the original phage display construction methods, this platform can prompt higher affinity and diversity for construction of scaffold library.
The plant homeodomain (PHD) finger domain, also called leukemia associated protein domain, was first identified in Arabidopsis thaliana HAT3.1, a protein involved in plant root development. It has been found as a kind of zinc binding domains in more than 600 eukaryotic proteins that involved in chromatin-mediated gene regulation. PHD finger motifs identified to date are between 50 and 100 residues in length and are characterized by a Cys4-His-Cys3 motif that share consensus sequences C-X1-2-C-X9-21-C-X2-4-C-X4-5-H-X2-C-X12-46-C-X2-C, where the 8 underlined cysteine residues coordinate two zinc ions. Structurally, PHD finger adopts a globular fold with no disulfide bonds which composed of a two-stranded β-sheet and an α-helix, and there are three loops formed at sequence regions X9-21, X4-5 and X12-46, respectively.
Based on its natural diversity and structural features, PHD finger motif has been examined as a scaffold for the presentation of selected binding functions. The PHD finger region consisting of secondary structures and the residues involved in coordinating the zinc-ions are very conserved among species. While, the relevant alignment of various PHD finger sequences that collected from different natural proteins has shown little conservation in the sequence of two PHD finger loops, which may contribute to the functional binding specificity of individual PHD finger. These two variable loops (at X9-21 and X12-46) seem to be tolerant to mutagenesis, expansion and loop grafting. Actually, by grafting the binding site for the co-repressor CtBP2 onto the PHD finger domain, an engineered PHD domain specifically binding CtBP2 has been achieved for GST pull-down assay or the yeast two-hybrid system. Therefore, it is possible to generate PHD finger scaffold libraries for engineering novel PHD fingers with new binding specificities and high affinity, which can be totally achieved by Creative Biolabs.
Creative Biolabs has developed the proprietary Hi-Affi™ phage library display platform to enhance the affinity and diversity of library construction. Our Hi-Affi™ platform combines phage display with trimer codon technology and NNK method to create a large repertoire of variants for selection of the best target-binder, enabling our experienced experts to construct mutant library with 100% precision and over 1010 diversity.
As an industry leader of scaffold protein research, Creative Biolabs has years of research and development experience in the field of scaffold library construction. Our scientists have successfully generated some 55 different kinds of scaffold libraries for our clients all over the world. Timely quotation, high quality service, fast delivery cycle and the most competitive price have been our commitment and guarantee to keep the leadership in the scaffold library construction market.
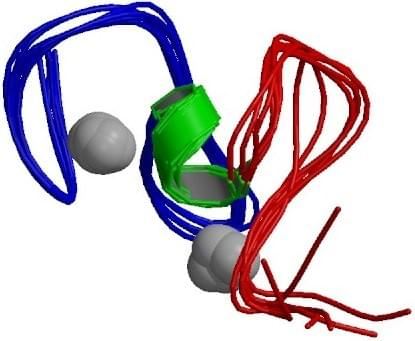 Fig. 1 Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. (PDB ID: 1F62)
Fig. 1 Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. (PDB ID: 1F62)
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.