While bacterial display has been increasingly used for screening antibody libraries, this technology has proven challenging in the screening for protein-specific antibody fragments. As a leader for bacterial display, Creative Biolabs has developed advanced anchored periplasmic expression (APEX) system to enable screening of antibody fragments. We provide several kinds of antibody libraries for bacterial display.
Bacterial display (also known as bacteria display or bacterial surface display) libraries contain billions of diverse molecules polypeptides on the surface of bacteria (for instance, on pili, flagella subunits or outer membrane proteins of Escherichia coli) to identify target peptides or proteins. Magnetic-activated cell sorting (MACS) or fluorescence-activated cell sorting (FACS) are two techniques used in bacterial display. These two techniques allow real-time analysis of displayed peptide properties, including target-binding affinity, specificity, and peptide stability to proteases during screening.
Anchored Periplasmic Expression (APEX) System for Antibody Fragments Screening
Antibodies engineered for intracellular function are required to have higher standards. They must not only have affinity for their target antigen but must also be soluble and correctly folded in the cytoplasm. Creative Biolabs provides APEX technique which enables the rapid isolation of target-specific antibodies without labor intensive screening and enables antigen-binding and proper cytoplasmic folding simultaneously. We have the ability to screen unique single-chain variable fragment (scFv) antibodies that have both high specificity and extremely high affinity, with Kd up to 10-12 for the target antigen.
The APEX technique displays a scFv library on the Escherichia coli inner membrane using twin-arginine translocation (Tat) pathway. Single-chain antibodies were anchored to the periplasmic face of the inner membrane via a fatty acylated lipoprotein sequence. The Tat pathway ensures that only soluble, well-folded proteins are transported out of the cytoplasm and displayed on the inner membrane for binding antigens.
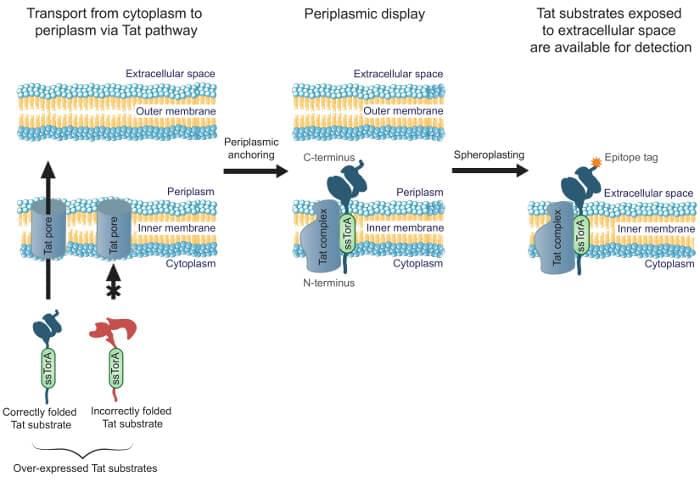 Figure 1. Tat inner-membrane display. (Moghaddam-Taaheri et al. 2016)
Figure 1. Tat inner-membrane display. (Moghaddam-Taaheri et al. 2016)
After scFv displayed on the periplasmic face of the inner membrane, we can remove the outer membrane using lysozyme. After this step, the scFvs are exposed and can be panned against fluorescently tagged antigens and screening with FACS to isolate scFvs that bind to the target antigen. An enzyme-linked immunosorbent assay (ELISA)-based secondary screen is used to identify the most promising scFvs for additional characterization. Antigen-binding and cytoplasmic solubility can be improved with rounds of mutagenesis and screening. The APEX system can also be used to express target antigens directly in conjunction with the library.
In addition to display scFv by bacterial display, we also have the ability to screen libraries of whole IgG antibodies (Fc binding protein) in the periplasm of E. coli. Besides, we can use multiple fluorescence parameters during screening by FACS, which allowing for screening for peptides and antibodies with well-defined specificity or cross-reactivity.
For more detailed information, please feel free to contact us or directly sent us an inquiry.
References
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.