Creative Biolabs offers customers a novel strategy for designing de novo protein libraries, the Binary Patterning of polar and nonpolar amino acids, to produce proteins with novel structures and functions for use in industry or medicine.
Protein structures have tolerance of amino acid substitutions: many different amino acids can encode the information necessary to produce a given three-dimensional structure. Taking advantage of this tolerance, we have employed a “Binary Patterning” strategy for protein design. Binary Patterning is based on the premise that the appropriate arrangement of polar and nonpolar residues can direct a polypeptide chain to fold into amphipathic elements of secondary structures, which can further anneal together to form a desired tertiary structure. This strategy incorporates both rational design and combinatorial diversity based on the “Binary Patterning” of polar and nonpolar amino acids. It is a potential tool for designing libraries of novel proteins.
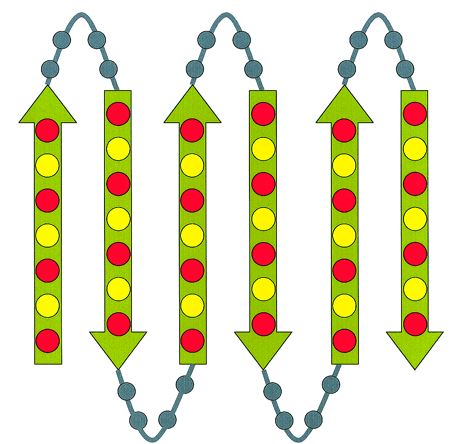
Figure 1. The designed binary patterning for a combinatorial library of de novo proteins. (Guofeng Xue et al., 2001)
In designing proteins with the binary pattern, the side chain identities can be varied extensively, enormous combinatorial diversity is possible, and degenerate codes can be used (i.e., NTN encodes nonpolar amino acids, and NAN encodes polar amino acids). Becauseα-helices have a repeating periodicity of 3.6 residues per turn, an amphipathic helix can be encoded by placing a nonpolar amino acid at every third or fourth position, with one face being polar and the opposite face nonpolar. The binary pattern will have to be adjusted for longer α-helices, to allow the 3.6 residues per turn periodicity to maintain the amphipathic nature the entire length of the helix. β-strands have an alternating periodicity of polar and nonpolar amino acids, causing one face of the strand to be polar and the opposite face to be nonpolar as well. To design turns, positional preference in natural proteins can be used as a reference (e.g., glycine in between helices), restriction sites need to be incorporated for easy gene assembly, and constant regions must be long enough for easy annealing. Besides, the protein length must be short enough to be error-free, long enough for proper folding, and resembling natural sequences. We can design your proteins using the binary method, with the length and key residues optimized for both stability and functionality.
Reference
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.