N-glycosylation Analysis of Proteins
N-linked glycosylation and O-linked glycosylation are both ubiquitous and essential post-translational modifications. N-linked glycosylation refers to the attachment of oligosaccharide chains to the nitrogen atom of asparagine residues in proteins. It begins in the ER with the en bloc transfer of a lipid-linked oligosaccharide (Glc3Man9GlcNAc2) to nascent polypeptides at the Asn-X-Ser/Thr consensus motif.
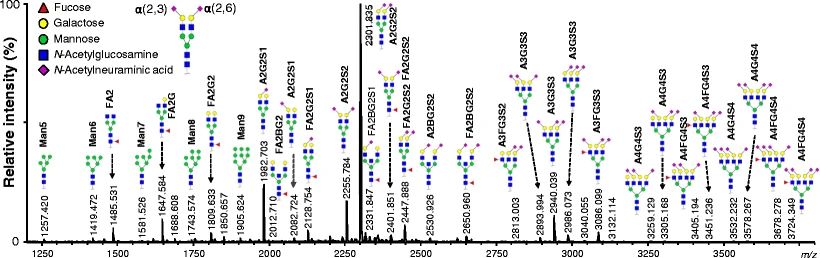 Fig.1 N-glycosylation landscape of human plasma proteins.1
Fig.1 N-glycosylation landscape of human plasma proteins.1
Process in the ER and Golgi
-
In the ER:
- Assembly: Carried out by ALG gene products onto Dolichol phosphate carriers.
- Trimming & Folding: α-glucosidases I and II remove terminal glucose, which facilitates protein folding by allowing interaction with calnexin/calreticulin chaperones.
- This chaperone cycle ensures proper folding, and misfolded proteins are retained or degraded.
-
In the Golgi:
- Processing into subtypes: Complex, hybrid, or high-mannose forms via trimming and extension of the core glycan (GlcNAc₂Man₃).
- Further additions include fucose, sialic acid, galactose, leading to structural heterogeneity critical for protein function and trafficking.
Biological Importance
- N-glycosylation directly stabilizes protein tertiary structure and indirectly aids through lectin-mediated chaperone recognition.
- Acts as a tag for the ER-associated degradation (ERAD) pathway if folding fails.
- Influences sorting and secretion of proteins, including immune receptors and enzymes.
Benefits of N-linked Glycosylation
| Functional Benefit | Mechanism/Impact |
|---|---|
| Protein folding & stability | Via ER chaperones (calnexin/calreticulin), stabilization of folding intermediates |
| Intracellular trafficking | Ensures ER-to-Golgi transition and efficient membrane targeting |
| Immunogenic modulation | Specific glycan motifs regulate immune recognition |
| Therapeutic half-life extension | Alters serum clearance rates of therapeutic proteins |
Step-by-Step Workflow for N-Linked Glycosylation Analysis
1. Protein Preparation & Enrichment
- Accepted samples include cell lysate, secreted proteins, purified recombinant glycoproteins.
- Enrichment is an optional but common step before analysis. We use lectin affinity (e.g., ConA for high mannose), hydrazide chemistry, or HILIC (hydrophilic interaction liquid chromatography) for glycopeptide/glycoprotein capture.
2. Glycan Release
- Enzymatic method is commonly used in this section while chemical method is rarely used due to harshness
- Use PNGase F to release N-glycans from Asn-X-Ser/Thr motifs.
- PNGase A can be used if α1,3-fucose is present (e.g., in plant glycoproteins).
3. Glycan Labeling (Optional for Enhanced Detection)
- Improves fluorescence and MS ionization.
-
Common fluorescent tags:
- 2-AB (2-aminobenzamide)
- Procainamide, 2-AA, or PA (pyridylamination)
4. Separation Techniques
A. Released Glycans
- HILIC-UPLC or CE with FLR detection for glycan profiling.
- MALDI-TOF-MS or LC-MS/MS for mass determination and structural info.
B. Glycopeptides
- Perform proteolysis (e.g., trypsin) on native or deglycosylated protein.
-
Analyze by LC-MS/MS for:
- Glycosite occupancy
- Glycan heterogeneity per site
- Peptide backbone info
5. Structure Elucidation
- Use exoglycosidase arrays or MS/MS fragmentation to determine branching, terminal residues (e.g., sialic acid, fucose), and linkage positions.
-
For deep analysis, combine:
- MSn (multiple-stage fragmentation)
- NMR spectroscopy (for linkage/conformation confirmation)
- Glycan sequencing tools
6. Data Analysis Tools
- Glycopeptide identification
- Predict glycan compositions from MS data
- Annotate and visualize glycan structures
- Quantitative MS analysis of glycopeptides
7. Quantification
- Relative: Based on peak areas from FLR or MS signal intensities.
- Absolute: Use glycan standards or stable isotope-labeled internal standards.
Analytical Techniques for N-Glycosylation
- HILIC-FLR-MS is widely used in industrial QC pipelines for its balance of throughput, resolution, and MS compatibility.
- LC-MS/MS of glycopeptides is the gold standard for site-specific glycosylation analysis.
- CE-LIF is a regulatory-approved method (e.g., for biosimilars), especially in Europe and Asia.
- MALDI-MS is often used as a screening tool or in early-stage glycan discovery.
- More techniques for N-glycosylation and their pros and cons are as followed:
| Technique | Analyte | Detection | Strengths | Limitations | Typical Applications |
|---|---|---|---|---|---|
| HILIC-FLR/UV-MS | Released glycans | Fluorescence/MS | High resolution, quantifiable, compatible with MS | Requires derivatization (e.g., 2-AB, RFMS); cannot localize site | Profiling glycan heterogeneity in mAbs and glycoproteins |
| MALDI-TOF-MS | Released glycans | MS only | Fast, sensitive, minimal sample needed | Limited quantitation; isomer resolution poor | Rapid glycan mass profiling; glycoform distribution |
| CE-LIF | Released glycans | Laser-induced FLR | High separation efficiency, low sample amount | Specialized equipment; limited structural info | High-throughput glycan profiling; QA/QC for biologics |
| LC-MS/MS (Bottom-up) | Glycopeptides | ESI-MS/MS | Site-specific glycosylation, glycan + peptide info | Complex data analysis; signal suppression from glycan moiety | Mapping glycosites, studying microheterogeneity |
| Top-down MS / Middle-down MS | Intact glycoproteins | ESI-MS | Intact glycoform distribution, charge envelope visualization | Requires ultrahigh-res instruments; lower sensitivity | Intact glycoprotein characterization; mAb Fc glycan profiling |
| Glycan Array (Lectin-based) | Native glycans | Fluorescence | Functional glycan-binding assay | Indirect; no structural info or quantification | Glycan-binding studies (e.g., Siglec or lectin profiling) |
| Exoglycosidase Digestion + MS | Released glycans/glycopeptide | MS/MS | Linkage, branching elucidation | Labor-intensive, sequential steps | Structural elucidation; verifying sialylation/fucosylation |
| NMR Spectroscopy | Pure glycans | NMR (1H, 13C) | Linkage and anomeric configuration; unambiguous structure | Requires large amount of purified glycans; low sensitivity | Confirming full glycan structure; reference standard creation |
What We Offer
N-linked glycosylation governs a wide array of biological processes, and its dysregulation underlies various human diseases—from congenital glycosylation disorders and autoimmune diseases like rheumatoid arthritis, to tumor progression and viral pathogenesis. At Creative Biolabs, we offer a full suite of customizable N-glycosylation research services to help you decipher glycan-related mechanisms and translate them into therapeutic opportunities.
✔ Glycosylation Profiling & Structural Characterization
Our high-resolution LC-MS/MS and HILIC-FLR platforms enable accurate mapping of N-glycan structures, glycosite occupancy, and branching patterns in both native and recombinant glycoproteins.
✔ Site-Specific Glycopeptide Analysis
We specialize in site-resolved glycoproteomics to uncover disease-specific glycan alterations—ideal for biomarker discovery in cancer and autoimmune diseases.
✔ Therapeutic Glycoprotein Engineering
Using glycoengineering strategies (e.g., CHO Lec mutants, FUT8 knockout, ENGase-mediated glycan remodeling), we help clients design recombinant antibodies, Fc-fusion proteins, or enzymes with optimized N-glycoforms for enhanced pharmacological performance.
✔ Anti-Glycan Immunotherapy Development
We offer custom services to generate anti-glycan antibodies, glycan-conjugated vaccines, and tumor-associated carbohydrate antigen (TACA) screening platforms—empowering glycan-targeted cancer immunotherapy.
FAQs
Reference:
- Clerc, Florent, et al. "Human plasma protein N-glycosylation." Glycoconjugate journal 33 (2016): 309-343. Distributed under Open Access license CC BY 4.0, without modification. https://doi.org/10.1007/s10719-015-9626-2