O-glycosylation Analysis of Proteins
Protein O-glycosylation—defined by the attachment of glycans to serine or threonine residues—represents a crucial post-translational modification (PTM) with broad implications for protein structure, function, and cellular signaling. Unlike N-linked glycosylation, which predominantly occurs in the ER and Golgi, O-linked glycosylation often takes place in the nucleus and cytoplasm and is highly dynamic and context-dependent. At Creative Biolabs, we specialize in high-resolution, site-specific protein O-glycosylation analysis services to support basic research and biopharmaceutical development.
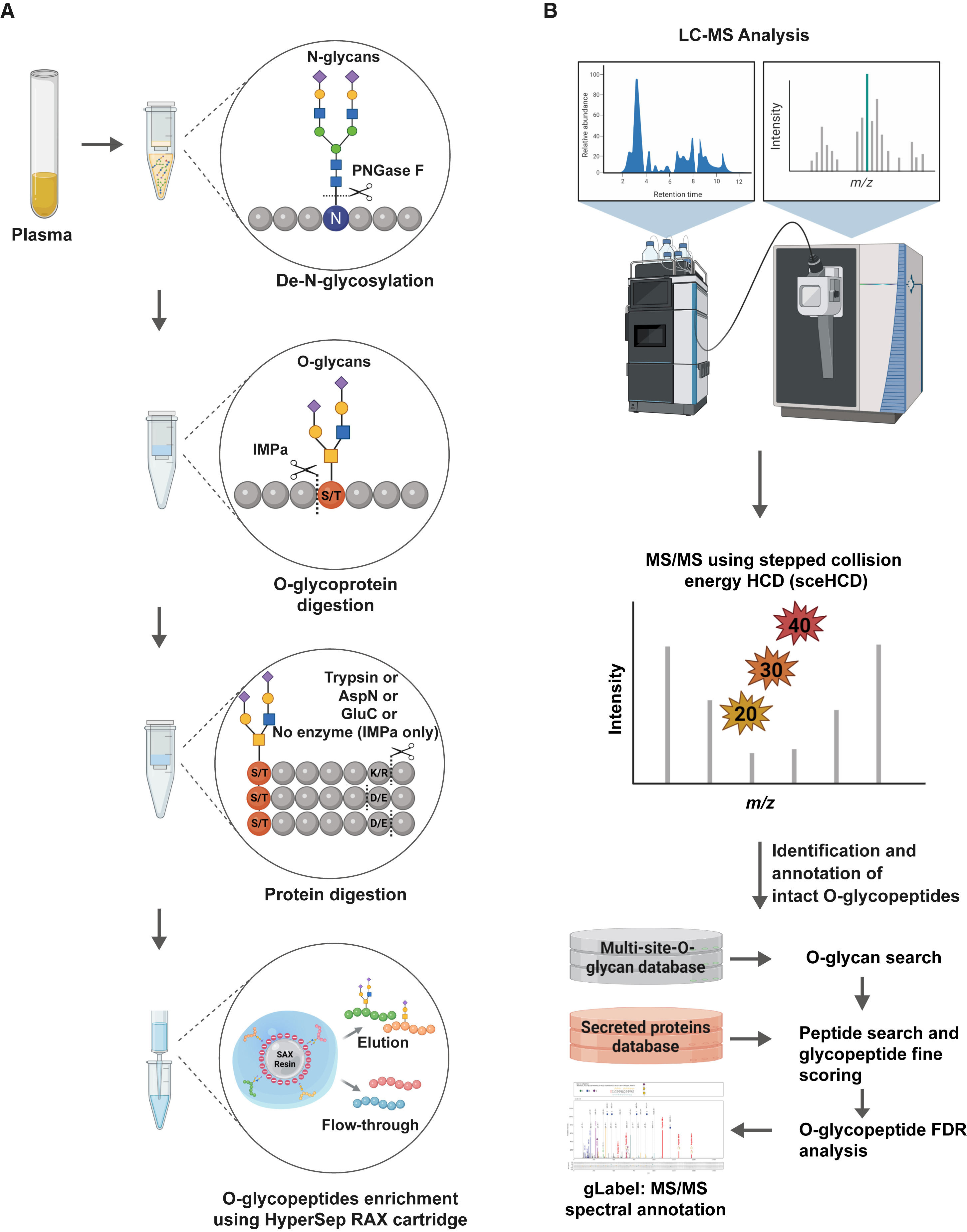 Fig.1 Integrated MS workflow for site-specific enzymatic O-glycoproteomics analysis.1
Fig.1 Integrated MS workflow for site-specific enzymatic O-glycoproteomics analysis.1
Types of O-Glycosylation
| O-Glycosylation Type | Linkage | Common Residues | Example Proteins | Functional Relevance |
|---|---|---|---|---|
| O-GalNAc | α-GalNAc-Ser/Thr | Mucins, Fc-fusion proteins | MUC1, MUC2 | Cancer, cell adhesion |
| O-GlcNAc | β-GlcNAc-Ser/Thr | Cytoplasmic/nuclear proteins | c-Myc, RNA Pol II | Phosphorylation crosstalk |
| O-fucose | Fuc-Thr | EGF-like domains | Notch1, Factor VII | Development, coagulation |
| O-mannose | Man-Ser/Thr | Brain proteins | α-dystroglycan | Muscular dystrophy |
| O-glucose | Glc-Thr | TSR domains | Thrombospondin-1 | Cell adhesion |
| O-glycosaminoglycan | Xylose-Ser | Proteoglycans | Decorin, Aggrecan | ECM organization |
O-GalNAc glycosylation, the most studied form, is catalyzed by polypeptide GalNAc-transferases (GALNTs), initiating mucin-type glycosylation in the Golgi apparatus. O-GlcNAcylation, in contrast, occurs in the cytoplasm and nucleus and is highly dynamic, interacting with phosphorylation pathways to regulate gene expression and protein stability.
Biological Roles of O-Glycosylation
- Protein Stability and Folding
O-glycans contribute to protein stabilization, enhance solubility, and modulate degradation. For example, O-GlcNAcylation of HGS accelerates its proteolytic turnover, while it stabilizes PPM1K in hepatocellular carcinoma. O-glycans can also act as folding tags, facilitating protein maturation via interactions with lectins and chaperones—though to a lesser extent than N-glycans.
- Cell Adhesion and Signaling
O-glycosylation influences intercellular adhesion through cadherins, selectins, and integrins. For instance, mucin-type O-glycans, especially sialylated and fucosylated forms, promote cell–cell and cell–matrix interactions during tumor progression.
- Phase Separation and LLPS
Emerging evidence links O-glycosylation with liquid-liquid phase separation (LLPS), influencing processes such as synaptic signaling and RNA translation. For example, O-GlcNAcylated SynGAP modulates LLPS behavior of PSD-95 complexes.
Analytical Methods for O-Glycosylation
Comprehensive O-glycosylation analysis requires multi-modal strategies combining glycoproteomics, glycomics, and imaging technologies.
| Technique | Applications | Strengths | Limitations |
|---|---|---|---|
| Mass Spectrometry (MS/MS) | Site-specific glycopeptide analysis | High sensitivity and resolution | Requires enrichment, poor ionization for some glycans |
| Capillary Gel Electrophoresis (CGE) | Structural profiling of oligosaccharides | High throughput, robust for small differences | Needs derivatization and fluorescent tagging |
| Lectin Microarrays | Glycan pattern recognition | Rapid screening, high throughput | Limited specificity, often semi-quantitative |
| Polyacrylamide Gel Electrophoresis | O-GlcNAc dynamics, protein resolution | Simple and economical | Not suitable for full structural elucidation |
| NMR Spectroscopy | Glycan conformation and stereochemistry | Label-free, non-destructive | Low sensitivity, large sample requirement |
| Imaging with Fluorescent Tags | Cell-surface glycan visualization | Spatial resolution, in situ detection | Dependent on dual-probe specificity |
Step-by-Step: How to Analyze O-Linked Glycosylation
Here's a step-by-step guide to O-glycosylation analysis:
1. Sample Preparation
- Protein source: Start with purified glycoproteins or complex mixtures (e.g., serum, cell lysates).
- Denaturation/reduction/alkylation: Essential for protein unfolding and optimal enzyme accessibility.
- Proteolytic digestion: Trypsin, Glu-C, or non-specific proteases (for better glycopeptide coverage).
- De-N-glycosylation (optional): Samples are de-N-glycosylated using PNGase F, avoiding interference of N-glycan.
2. Glycan or Glycopeptide Enrichment (Optional)
- Lectin affinity chromatography: For Jacalin (binds core 1 Galβ1-3GalNAc), VVA (binds Tn antigen).
- HILIC (Hydrophilic Interaction Liquid Chromatography): Enriches hydrophilic glycopeptides—widely used before LC-MS.
- Strong anion exchange (SAX): Used for sialylated glycopeptides.
3. Release of O-Glycans (if analyzing free glycans)
Unlike N-glycans (which can be enzymatically removed by PNGase F), no universal enzyme exists for O-glycans. Instead:
- Reductive β-elimination: Releases O-glycans under alkaline conditions (NaOH/NaBH₄); destroys peptide backbone
- Non-reductive β-elimination: Preserves glycan reducing end for MS; compatible with fluorescent tagging
Caution: Harsh conditions can lead to peeling (sequential loss of sugars).
4. Labeling (Optional, for Glycomics)
- Fluorescent tags (e.g., 2-AB, 2-AA, RapiFluor-MS) improve detection and MS signal.
- Often used in conjunction with HILIC-UHPLC or CE-LIF.
5. Mass Spectrometry (MS) Analysis
a) Released O-Glycans
- MALDI-TOF-MS or ESI-MS (after labeling or permethylation)
- Provides composition but not linkage or position
b) Glycopeptides
- LC-MS/MS using HCD, ETD, or EThcD fragmentation
- Allows simultaneous peptide sequence and glycan composition identification
- ETD is particularly useful for site localization
6. O-Glycosite Mapping
- Use ETD/EThcD fragmentation of glycopeptides
- Software tools are commonly used.
- Validate with synthetic standards if available
7. Quantitation (Optional)
- Label-free quantitation: Based on MS peak area
- Isobaric tags (e.g., TMT): Less common for glycopeptides due to complexity
- Metabolic labeling: Possible in cultured cells using sugar analogs (e.g., Ac4GalNAz)
Clinical Implications
O-Glycosylation and Cancer
Aberrant O-glycosylation patterns—particularly truncated O-glycans like Tn and sTn antigens—are widely observed in carcinomas. For instance, upregulation of GALNT7 enhances O-glycosylation in prostate cancer, contributing to tumor proliferation. Mucin-type O-glycosylation, notably in colorectal and pancreatic cancers, correlates with immune evasion and metastatic potential. These patterns offer biomarker potential for early-stage detection and treatment stratification.
Glycoengineering and Therapeutics
Advances in glycoengineering platforms, including selective editing of O- and N-glycan structures on living cells, have enabled functional studies and improved biotherapeutics. For example, receptor O-glycosylation was found to regulate dimerization and internalization of opioid receptor OPRD1, independent of N-glycosylation status.
A Case Example: O-Glycosylation in CNS and Autoimmunity
Recent research links O-GlcNAcylation dysregulation to neurodegenerative diseases, including Alzheimer's and Parkinson's. Alterations in OGT/OGA activity modulate levels of α-synuclein and tau protein aggregation. In immune responses, cell surface O-glycans modulate Siglec interactions, influencing inflammation, monocyte adhesion, and immune checkpoint signaling.
Future Insights and Our Solutions
O-glycosylation's complexity has historically limited our understanding. However, innovations are driving breakthroughs in diagnostics and targeted therapies:
- Single-cell glycoproteomics
- Site-specific gene editing of GALNTs
- Glycoprotein visualization via ARPLA imaging
- Machine learning-assisted glycosite prediction
O-linked glycosylation is a structurally rich and functionally pivotal PTM with vast implications in biology and medicine. From Fc-fusion biologics to oncogenic mucins, understanding and accurately profiling O-glycosylated proteins is no longer optional—it's essential. At Creative Biolabs, we are committed to advancing glycoproteomics by offering precise, custom-tailored O-glycosylation analysis services to support:
- Biologic drug development
- Cancer glycomarker discovery
- Structural-functional glycoprotein analysis
Let Creative Biolabs be your partner in unraveling glycan complexity. Our integrated platform of MS, CE, enzymatic mapping, and custom analytics ensures a deep, actionable understanding of your protein O-glycosylation landscape.
Reference:
- Kang, Taewook, et al. "Global O-glycoproteome enrichment and analysis enabled by a combinatorial enzymatic workflow." Cell Reports Methods 4.4 (2024). Distributed under Open Access license CC BY 4.0, without modification. https://doi.org/10.1016/j.crmeth.2024.100744