It is essential for research to keep pace with the constant evolution of pathogens in order to discover the determinants of drug resistance, immune evasion, and virulence in the study of infectious diseases. Transcriptomics research can help in the study of infectious diseases by analyzing changes in gene expression profiles to discover new therapeutic targets. Creative Biolabs can provide a full range of transcriptomics services to assist clients in accelerating research whether it is for vaccine development, antibiotic resistance, new viruses, or immune evasion.
Transcriptome sequencing is effective in analyzing global gene expression levels in infectious diseases and analyzing transcriptomes in depth, discovering novel transcripts and isoforms, as well as assembling previously unstudied transcriptomes to identify possible therapeutic targets. These analyses are involved as follows.
Step 1. Data preprocessing
The raw RNA sequencing data are preprocessed using standardized analysis. First, quality control of the raw RNA-seq sequences is performed to remove low-quality sequences and splice sequences. Then, the eluted sequences are aligned to the reference genome. Finally, estimate gene expression based on the gene-encoded information.
Step 2. Differential gene analysis
Calculate the expression of different genes in the sample. This analysis calculates the log2 fold change and p-value of the gene. log2 fold change value indicates the degree of difference in the expression of the gene in the two sets of samples, while the p-value represents whether the difference is significant or not. Genes with a p-value less than 0.01 are defined as differentially expressed genes.
Step 3. GO/KEGG enrichment analysis
GO analysis is able to annotate and categorize the biological functions of differentially expressed genes at different levels, such as cellular and molecular, etc. KEGG analysis is able to annotate the signaling pathways and metabolic pathways of differentially expressed genes. GO/KEGG results with a p-value less than 0.01 are considered significant.
Step 4. Screening for therapeutic targets
Among all differentially expressed genes, those with potential roles in the disease treatment process, such as those related to immune response, inflammatory response, etc., are selected first. Then, based on the results of GO/KEGG analysis, those functional items related to infectious diseases are selected, and genes with significant differential expression are further selected. These genes are considered as potential therapeutic targets.
Various types of samples such as total RNA, mRNA, ds-cDNA, and cultured cells/cell sediment are acceptable.
Sequencing Type
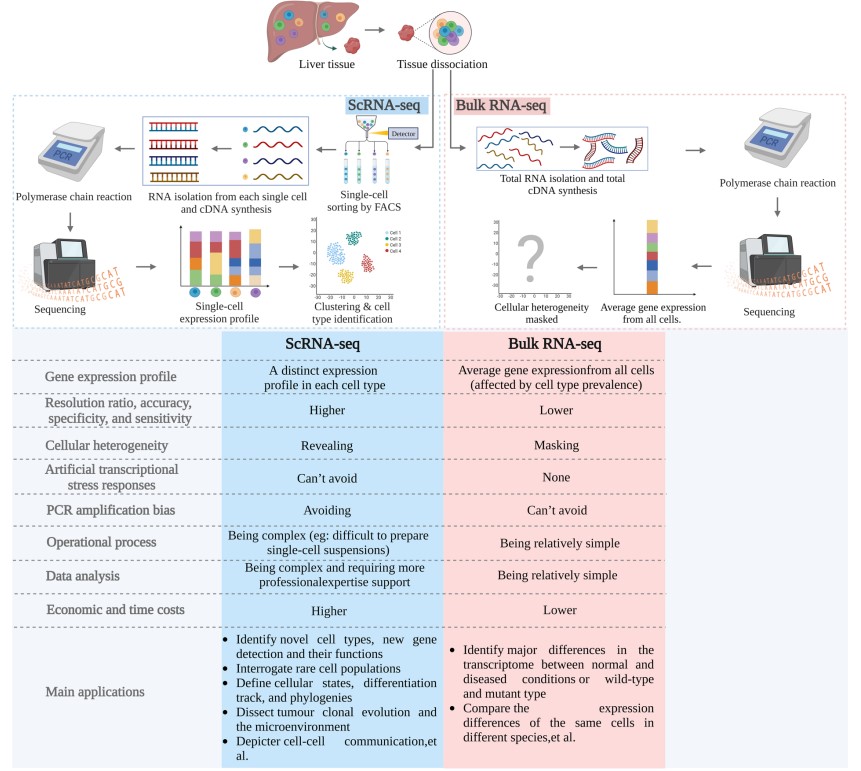 Fig. 1 Comparison of single-cell RNA sequencing and bulk RNA sequencing.1
Fig. 1 Comparison of single-cell RNA sequencing and bulk RNA sequencing.1
Starting from experimental design, and standardized experimental procedures to professional bioinformatics analysis, Creative Biolabs aims to provide transcriptome analysis solutions for your target discovery in infectious disease research. Please contact us if you are interested and wish to learn more.
Reference: