Creative Biolabs offers our expertise in DOGS for protein engineering. We work closely with our customers to design each project and meet your individual requirements.
Many different types of DNA Shuffling techniques have been used to improve the protein characteristics. This is done by the production of chimeric gene products based on a collection of mutants of the same gene or related families of genes. For example, error-prone PCR has been normally used, alongside misincorporation mutagenesis and subsequent gene shuffling to generate mutant genes that encode protein products of limited variation in sequence space; the identities of the protein products can only be defined via extensive screening. Another gene shuffling method is done after DNase I fragmentation of related genes (family shuffling); however, this method has an overwhelming percentage of wild-type parental sequences in the final shuffling products.
In order to solve the issue of persisting “parental” genes, DOGS is carefully designed. It is a procedure for DNA shuffling using degenerate primers, and enables shuffling of genes that differ widely in sequence similarity and G/C content, reducing significantly the regeneration of wild-type genes. In fact, this method can control the relative levels of recombination between the shuffled genes and the unshuffled parental genes. The primer extension conditions may be further modified to selectively produce progeny genes toward any one (or more) of the parental input genes. Additionally, it avoids the use of endonucleases for gene fragmentation before shuffling.
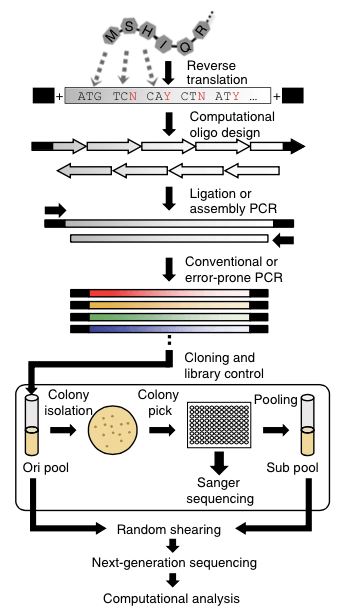
Figure 1. An illustration on the general scheme of gene library construction leading to the next-generation sequencing and computational analysis (Namjin Cho et al., 2014)
Modification of consensus–degenerate hybrid oligonucleotide primer (CODEHOP) design is done for DOGS. CODEHOP primers have a relatively short 3′degenerate end and a 5′nondegenerate consensus clamp: this design reduces the total number of individual primers needed, allows hybridization to be stabilized by the 5′ nondegenerate consensus clamp, and ensures the mismatches between the primer and the target to be less disruptive to priming of polymerase extension. Sequence similarity of the primers can improve the amplification and extension of primed PCR products. Modified CODEHOP technique uses perfectly complementary degenerate- end (CDE) primers, each of which contains a nondegenerate core flanked by both 5′ and 3′ degenerate ends. The corresponding coding sequence of one gene determines the nondegenerate core of an CDE primer, forming chimeric fragments that retain the parental sequence at the points of segment overlap.
Therefore, good primer design is the key of the DOGS procedure. We can work on your protein engineering projects with optimally designed primers for largest gene variations and lowest parental gene retainment.
Reference
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.