Biomarker Identification by ORFeome Phage Display
The identification of biomarkers from pathogens is a prerequisite for the development of vaccines and diagnostic assays. Creative Biolabs developed an extensive and comprehensive strategy, named ORFeome phage display, for the identification of biomarkers related to pathogens. We apply this technology to identify new biomarkers by using bacterial genome libraries. We are proud to introduce this service for new biomarkers identification to all our global customers.
Antibodies have been widely used in immunological tests for specific detection and identification of pathogens. The availability of monoclonal antibodies (mAbs) against bacterial surface antigens not only allows the development of detection assays but also provide a powerful tool for the study of bacterial protein structures and functions. Since mAbs recognize exclusive epitopes in the antigen, they can be used to identify new proteins that would be important in the bacterial pathogenesis, survival, or adaptation to the environment. Moreover, mAbs offer a highly reproducible reagent that can be produced in unlimited amounts. For example, Listeria is one of these pathogens. L. monocytogenes is the pathogen that predominantly infects humans resulting in severe infection in the elderly, cancer patients, HIV patients, pregnant women, and their fetuses or infants.
Through years' research, many groups have focused efforts on the production of mAbs specific to this genus with variable species specificity with unknown or uncharacterized antigen targets. At the same time, attempts with variable success were also made to develop L. monocytogenes-specific antibodies using specific target antigens. Although most of these works focused on the detection of the pathogenic species, non-pathogenic Listeria should be considered. For the reason that non-pathogenic Listeria grows faster, false-negative results may increase as well. Thus, the identification of new biomarkers that allow the detection of both pathogenic and non-pathogenic species is of high value for the development of detection methods.
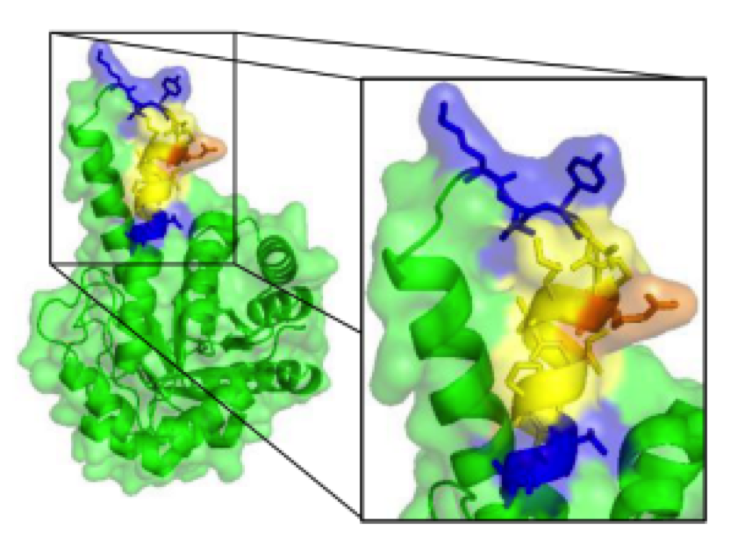 Fig.1 The position of the epitope in fructose-1,6-bisphosphate aldolase (FBA) structure shows it is a loop-to-helix transition, which has many amino acid side chains exposed on the protein surface.1
Fig.1 The position of the epitope in fructose-1,6-bisphosphate aldolase (FBA) structure shows it is a loop-to-helix transition, which has many amino acid side chains exposed on the protein surface.1
Creative Biolabs has developed this cutting-edge ORFeome display technology for the identification of new biomarkers of pathogens. The process includes (1) construction of an antigen library by using the genomic DNA of the target pathogen, (2) antigen panning, (3) ELISA for screening, (4) peptide membrane for the epitope mapping and (5) sequence and structure analysis. Through this method, our scientists have identified many new biomarkers for different pathogens.
Key advantages including but not limited to:
- Using the genomic DNA
- Extensive and comprehensive panning
- Accurate epitope
- Advanced phage display platform
- Identification of biomarkers for both pathogenic and non-pathogenic species
In the field of phage display technology, Creative Biolabs has extensive expertise, which has been accumulated through our over a decade of experience. We are pleased to provide this ORFeome phage display technology for the identification of new biomarkers of pathogens for all our clients.
For more detailed information, please feel free to contact us or directly sent us an inquiry.
Reference
- Mendonca, Marcelo, et al. "Fructose 1, 6-bisphosphate aldolase, a novel immunogenic surface protein on Listeria species." PloS one 11.8 (2016): e0160544. Distributed under Open Access license CC BY 4.0, without modification.
For Research Use Only.
