Pathogenicity Island (PAI) Identification
One of the important applications of bacterial genome analysis is the identification of pathogenicity islands (PAIs) namely virulence genes, which can provide insights into the pathogenesis of bacterial pathogens and promise antibacterial drug targets. Creative Biolabs has developed an excellent genomic sequencing and analysis platform to identify PAIs for further antibacterial drug targets identification.
PAIs (Figure 1) are present in the genomes of pathogenic organisms but absent from the genomes of nonpathogenic organisms of the same or closely related species. It is due to the PAIs were originally transferred from other pathogenic bacteria, plasmids or phages, and their sequences encode one or more virulence factors, such as adherence factors, toxins, invasions, and modulins. PAIs occupy relatively large genomic regions covering 10-200 kb. Identifying PAIs according to their genome signature includes:
G+C Content and GC-Skew
The G+C (%) contents in PAIs are often different from that of the host organisms. For example, the G+C content of the uropathogenic E. coli core genomes was 51%, while the G+C content was 41% in PAI I, II, IV, and V.
Codon Usage
Generally speaking, each genome has its preferred codon usage, and thus the codon usage in a genome region will be significantly different than the rest of host genome if this region was transferred from outside.
K-Mer Frequency
The measurements of dinucleotides or high-order oligonucleotide frequencies have been increasingly used. Theoretically, the higher-order measurement used, the more accurate to differentiate two genomes.
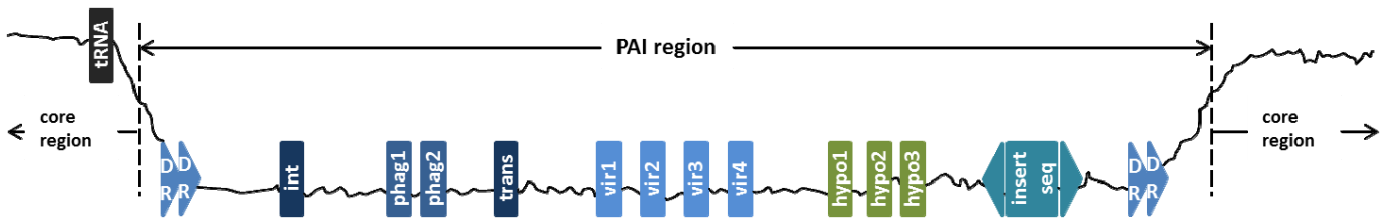 Figure 1. A schematic view of a pathogenicity island with associated features. (Dongsheng Che et.al. 2014)
Figure 1. A schematic view of a pathogenicity island with associated features. (Dongsheng Che et.al. 2014)
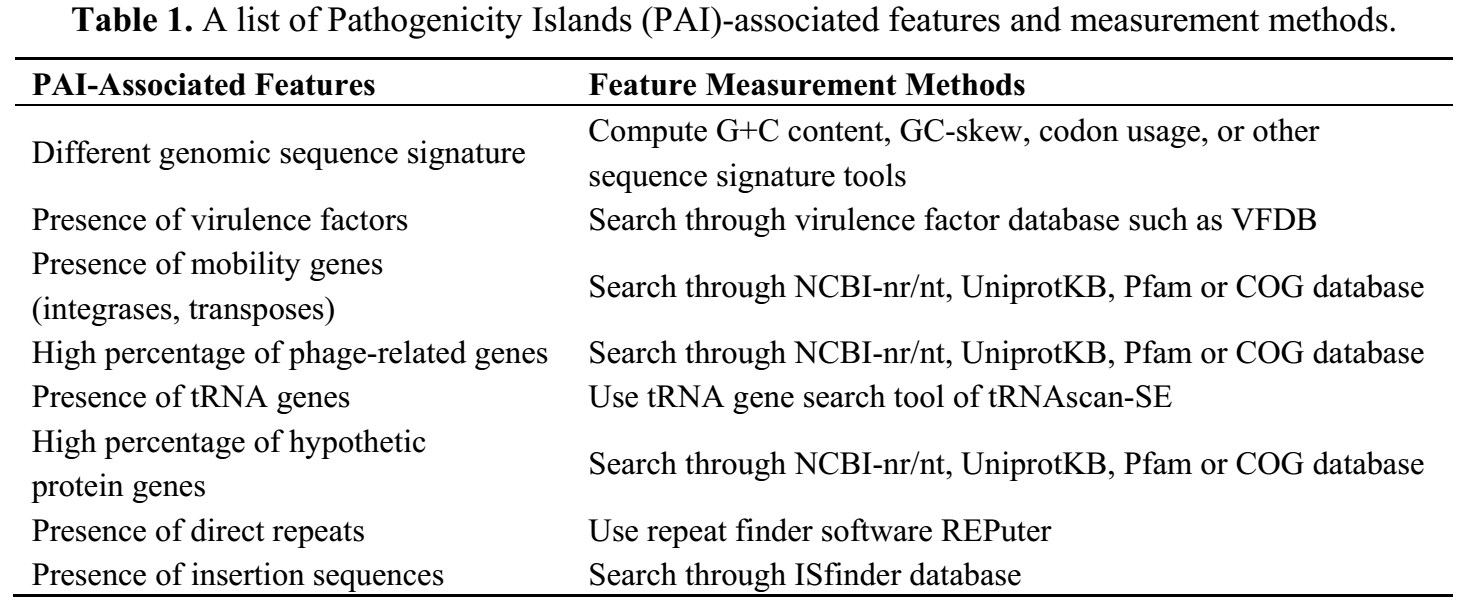
There are several ways of identifying PAIs or virulence genes according to their sequence features (Table 1). One method involves searching for regions with sequence signatures that are different from other parts of the genomic sequence. The second method for identification of PAIs is through comparative genomics analysis of closely related genomes or different genomes of species but cause similar infections. New virulence genes can also be identified by finding genes co-regulated with known virulence genes.
Table 1. A list of Pathogenicity Islands (PAI)-associated features and measurement methods. (Donkor 2013)
Comparative Genomics-Based Approach
Comparative genomics-based approach compares the incongruence of the gene tree versus its associated species tree. This approach consists of three general steps: collecting all genome sequences from closely related species for a query genome; aligning these genome sequences together; and then considering those gene segments present in the query genome but not present in others to be islands.
Sequence Composition Based Approach
All genomic regions inside the host genome are supposed to share same genomic signature. If a piece of genomic sequence has been detected with different gene signature or contents, it is highly recommended that this region is horizontally transferred from other sources. Creative Biolabs can identify special signatures according to G+C content, dinucleotide frequencies, codon usage, mobility genes, tRNA genes, and flanking direct repeats. In the case of PAIs, the region also contains virulence factor genes.
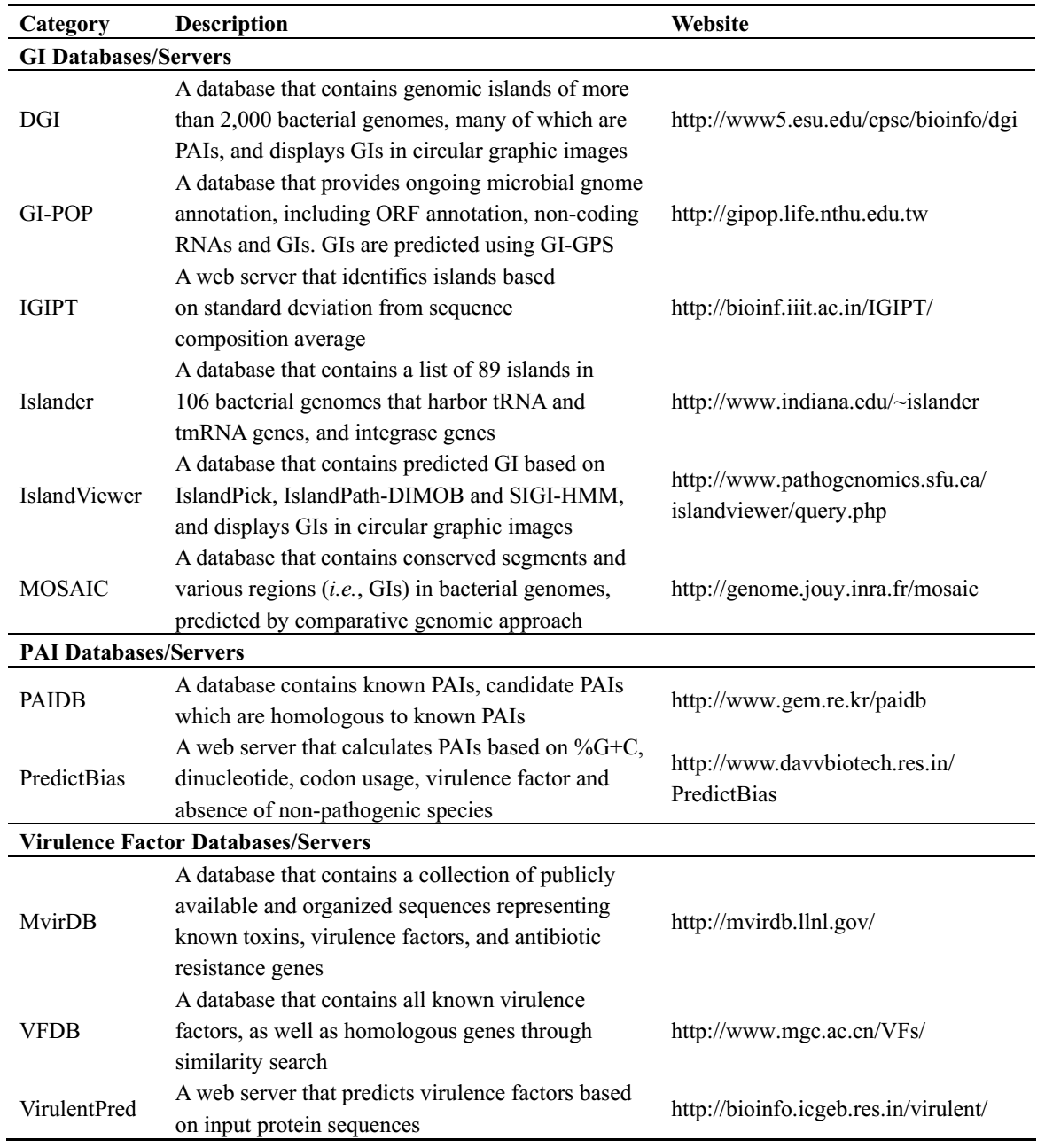
Other than above ways of identifying PAIs or virulence genes, there are a number of PAI related databases and online resources that can be accessed directly (Table 2). Creative Biolabs provides genomic sequencing and analysis platform to identify PAIs for further antibacterial drug targets identification.
Table 2. The summary of public island databases and web resources. (Che et.al. 2014)
For more detailed information, please feel free to contact us or directly sent us an inquiry.
References
- Donkor E S. (2013). “Sequencing of bacterial genomes: principles and insights into pathogenesis and development of antibiotics”. Genes 2013, 4:556-572
- Che D, Hasan M S and Chen B. (2014). “Identifying pathogenicity islands in bacterial pathogenomics using computational approaches”. Pathogens 2014, 3:36-56
For Research Use Only.
