Four-Dimensional Exosomal Proteomic Detection Service
Overview Services Features FAQs
Creative Biolabs provides proteomics services for 4D exosomes to facilitate proteomics and high-throughput modifications in micro samples and large sample groups.
Overview of Four-Dimensional Exosomal Proteomic
Determining the physicochemical characteristics of the tested sample ions is the fundamental idea behind mass spectrometry (MS) proteomics. This allows for the acquisition of both qualitative and quantitative results based on sample mass spectra and associated data. In order to identify and measure proteins, traditional proteomics mass spectrometry (MS) typically uses the three dimensions of retention time, m/z, and ion intensity, or 3D proteomics. By adding a fourth dimension, ion mobility, to 3D proteomics-primarily based on the section and shape of the ions to be separated-4D proteomics makes it possible to distinguish between and identify low-abundance protein signals.
4D Proteomics Assay Service at Creative Biolabs
At Creative Biolabs, we offer 4D proteomics assays for exosomes from many types of sample sources, such as Co-IP samples, tissue samples, and protein samples.
-
Co-IP samples have a protein requirement of around 2-5ug, no detergent (e.g. SDS), and a high oxygen concentration. Or the beads with the protein samples can be sent to us directly, and then we will undertake the subsequent experiments.
-
If tissue samples are provided, please send the tissue samples in dry ice conditions.
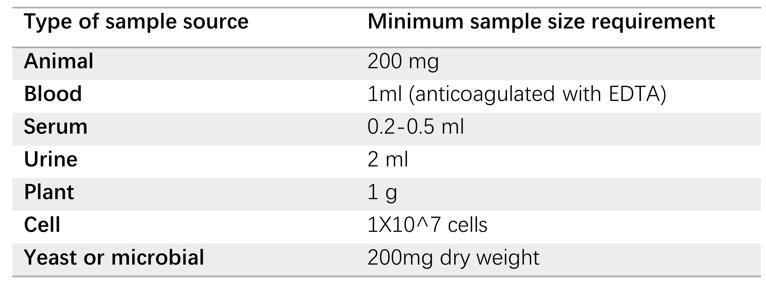 Fig.1 Tissue sample requirements.
Fig.1 Tissue sample requirements.
-
Protein samples with a total protein of 50ug or more. For protein extraction, ordinary tissue, and cell lysis solution can be used.
-
Sample transportation: Use plenty of dry ice and ship fresh specimens as soon as feasible to avoid the likelihood of sample degradation during transportation.
A detailed report on the 4D Exosomal Proteomic service will be provided at Creative Biolabs, which includes
-
The experimental procedures.
-
Relevant experimental parameters.
-
MS images.
-
Raw data.
-
Proteomics analysis results.
Features
-
High Sensitivity Detection: Our service utilizes state-of-the-art techniques to detect exosomal proteins with high sensitivity, enabling the identification of low abundance proteins and rare biomarkers.
-
Multiplex Analysis: With our four-dimensional approach, we can simultaneously analyze multiple aspects of exosomal proteomes including protein identification, quantification, post-translational modifications, and protein-protein interactions.
-
Rapid Turnaround Time: Our streamlined workflow allows for quick turnaround times, so you can get your results faster and accelerate your research progress.
-
Comprehensive Reporting: We provide detailed reports with comprehensive analysis results, including data visualization and interpretation, to help you make informed decisions for your research.
FAQs
Q: What types of samples can be analyzed using your service?
A: We accept various sample types including cell culture supernatants, plasma, serum, urine, and other bodily fluids and isolated exosome samples.
Q: How much sample is required for analysis?
A: We typically require a minimum of 10 µg of exosomal protein for analysis. However, the precise quantity could change based on the particular demands of your project.
Q: Can you analyze exosomal proteins from clinical samples?
A: Yes, we have the capability to analyze exosomal proteins from clinical samples, enabling translational research and biomarker discovery studies.
Creative Biolabs provides a 4D exosome proteomics service, which optimizes the detection cycle time, depth of identification, and high throughput performance based on traditional proteomic identification to facilitate the in-depth proteomics of exosome-related complex samples. Please feel free to contact us.
For Research Use Only. Cannot be used by patients.
Related Services:

 Fig.1 Tissue sample requirements.
Fig.1 Tissue sample requirements.