Biotin Modification Service
Introduction
Biotin, a water-soluble vitamin B7, is a high-affinity ligand for streptavidin, ideal for oligonucleotide modification due to its small size and biocompatibility, which don't compromise target-binding specificity. Biotinylation enhances oligonucleotide stability by improving nuclease resistance, facilitates cellular uptake by altering membrane charge interactions, enables precise intracellular tracking via streptavidin-based fluorescence or electron microscopy, and supports targeted drug delivery through integration with streptavidin-conjugated carriers. At Creative Biolabs, we offer end-to-end solutions: custom sequence optimization for specificity, high-precision synthesis with TEG-biotin or terminal modifications, and rigorous HPLC/MS validation, stability profiling, and functional assays to ensure batch consistency, helping harness biotinylated oligonucleotides for gene editing, diagnostics, and therapeutics.
[Contact Our Experts for Tailored Solutions]
Biotin
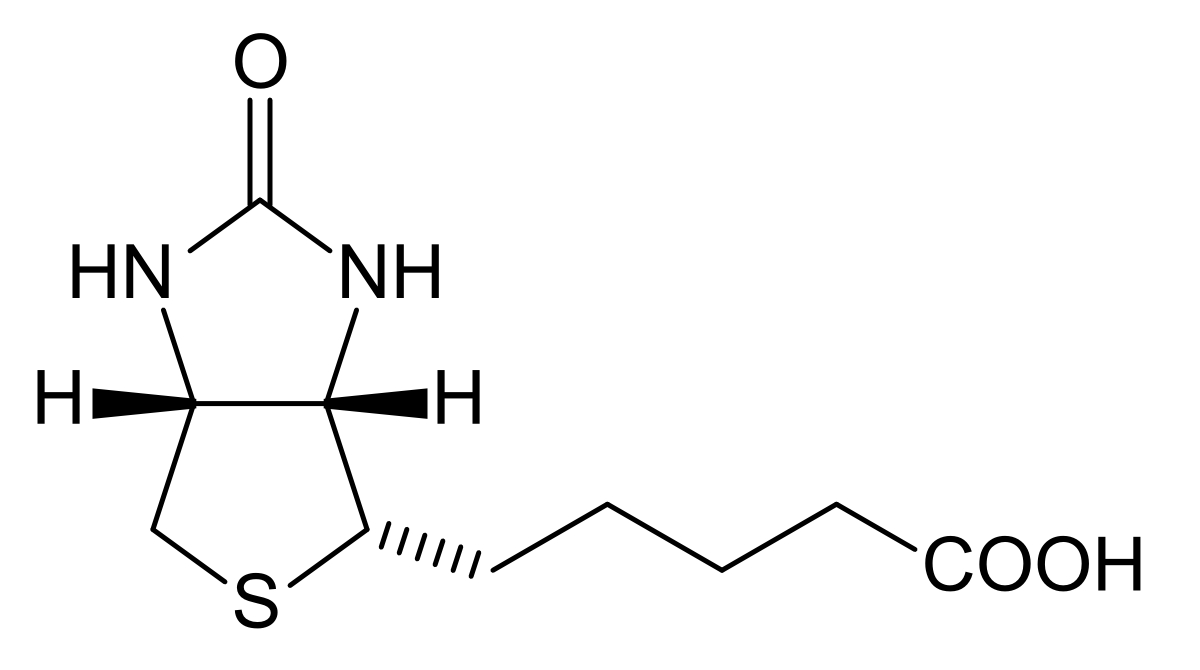 Fig.1 Chemical structure of biotin (vitamin H/B7).Distributed under public domain, from Wiki, without modification.
Fig.1 Chemical structure of biotin (vitamin H/B7).Distributed under public domain, from Wiki, without modification.
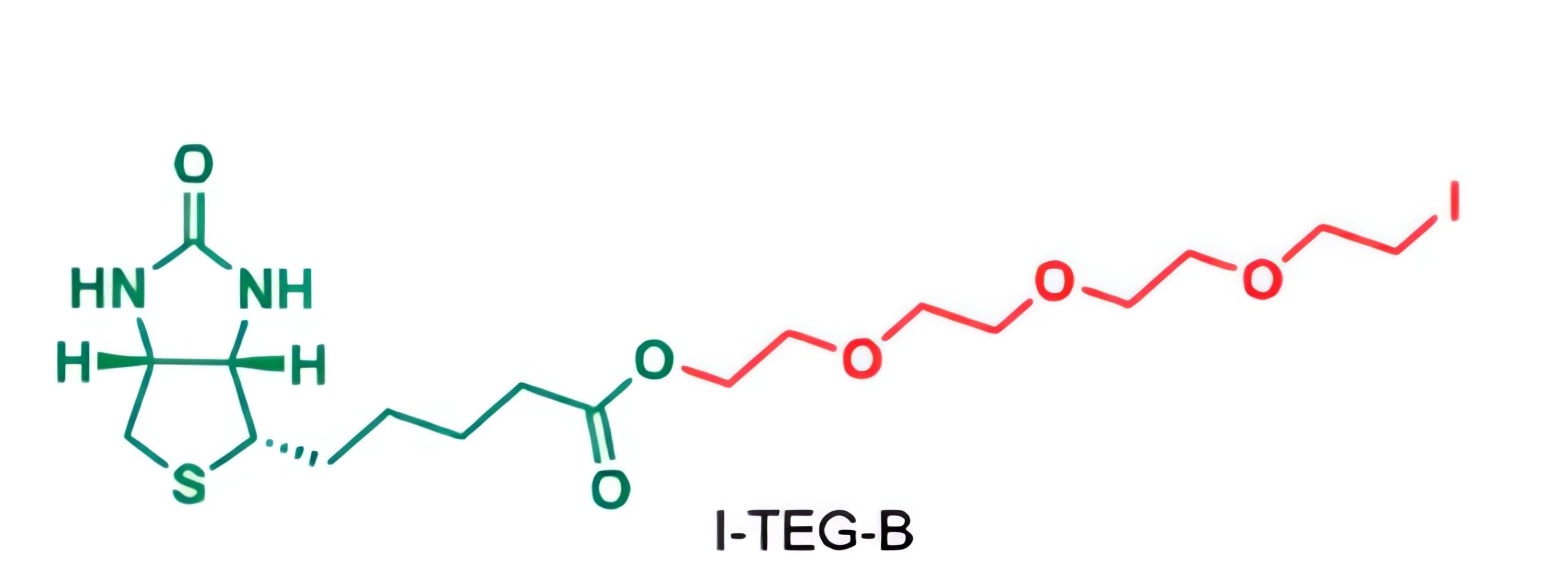 Fig.2 The esterification reaction of biotin with TEG diiodide (I-TEG-I) yields biotin-TEG-iodide (I-TEG-B).1
Fig.2 The esterification reaction of biotin with TEG diiodide (I-TEG-I) yields biotin-TEG-iodide (I-TEG-B).1
| Classification Criteria | Biotin Type | Characteristics | Application Scenarios |
|---|---|---|---|
| Linker Arm Length | Short-chain biotin (e.g., Biotin-TEG) | Linker contains 3–6 atoms; low steric hindrance; minimal impact on hybridization. | Scenarios requiring preservation of oligonucleotide conformation (e.g., qPCR probes). |
| Long-chain biotin (e.g., Biotin-dT, adipate- or PEG-chain derivatives) | Linker contains 10–16 atoms; reduces spatial interference; enhances streptavidin binding. | Solid-phase capture, magnetic bead separation, or detection in complex systems. | |
| Modification Position | 5'-end biotin modification | Linked via phosphodiester bond or amino linker; commonly used modification. | Oligonucleotide immobilization, pull-down assays for capturing target nucleic acids/proteins. |
| 3'-end biotin modification | Requires specialized synthesis (e.g., 3'-biotinylated nucleotide monomers). | Avoids interference with 5' primer-binding regions; suitable for PCR with 5'-end function preservation. | |
| Internal biotin modification | Introduced via side-chain modification of internal nucleotides (e.g., dT, as in Biotin-dT). | Complex designs requiring multi-labeling or site-specific internal biotinylation. | |
| Commercial Models | Biotin-TEG | Contains triethylene glycol linker; high water solubility. | Widely used for 5'/3'-end modifications. |
| Biotin-dT | Commonly used for internal modification; biotin linked to dT at position 5 without affecting base pairing. | Internal biotin modification. | |
| Biotin-SS-TEG | Contains disulfide bond; cleavable biotin. | Experiments requiring elution of biotinylated oligonucleotides. | |
| Biotin-11-dUTP | Enzymatically incorporated for DNA biotinylation; spacer arm contains 11 atoms. | Enzymatic biotinylation of oligonucleotides. |
Advantages
Efficient Separation and Purification
Biotinylated oligonucleotides and their bound targets (e.g., complementary nucleic acids, proteins) can be rapidly captured using streptavidin-coated magnetic beads, resins, or plates, achieving high-purity isolation (≥95% purity).
High-Sensitivity Detection
Combined with fluorescent or enzyme-labeled streptavidin, biotinylated oligonucleotides enable detectable signals for hybridization assays or quantitative analysis, with a detection limit as low as picomolar levels.
Stable Immobilization
Biotinylated oligonucleotides can be stably anchored onto streptavidin-modified solid supports (e.g., chips, electrodes, microspheres) for biosensor or nucleic acid array fabrication, with binding stability unaffected by pH, temperature, or ionic strength.
Minimal Interference
Biotin attachment via a linker arm minimally impacts oligonucleotide base pairing or secondary structure, ensuring uncompromised hybridization efficiency with targets.
Multifunctional Compatibility
Biotinylated oligonucleotides can be combined with other modifications (e.g., fluorescent labels, phosphorothioation) to enable dual functionalities such as "capture plus detection" or "targeting plus labeling."
Design
Selection of Modification Position
- 5' end modification: The most commonly used, with minimal impact on oligonucleotide hybridization. It is suitable for immobilization or as a marker for primers.
- 3' end modification: Care must be taken to avoid interfering with DNA polymerase extension. It is ideal for blocking the 3' end to prevent degradation.
- Internal modification: Non-critical sequence positions should be chosen to avoid disrupting base pairing. Modified dT is preferred, as its interference with base pairing is less than that of modified A/C/G.
Matching of Linker Arm Length
- Short-chain linker arms (3-6 atoms): These have low steric hindrance and are suitable for scenarios requiring preservation of oligonucleotide conformation. However, their proximity to the oligonucleotide may reduce binding efficiency with streptavidin.
- Long-chain linker arms: These reduce spatial interference and enhance binding with streptavidin, making them suitable for capture in complex systems. They may, however, increase non-specific binding.
Control of Biotin Quantity
- Single modification is preferred: Excessive biotin may cause streptavidin bridging effects, which can reduce separation efficiency.
- Special requirements: If enhanced binding is needed, two biotins can be introduced at both ends or in spacer sequences, but verification is required to ensure no impact on oligonucleotide function.
Workflow
-
Sequence & Modification Design Optimization
Based on the target sequence's GC content, secondary structure, and application scenarios (e.g., detection, targeted delivery), determine the biotin modification position (5'-end, 3'-end, or internal). Prioritize sites with minimal interference on hybridization: 5'-end modifications are ideal for immobilization, while internal modifications prefer non-critical sequences using modified dT.
Match linker arm length to needs: short-chain linkers (3-6 atoms) suit scenarios requiring preserved oligonucleotide conformation, while long-chain linkers (10-16 atoms) enable efficient capture in complex systems.
Control biotin quantity, with single modification as the standard to avoid streptavidin bridging from excess biotin. Dual modifications may be designed for special needs but require validation of their impact on oligonucleotide function.
-
Precision Synthesis
Use automated solid-phase synthesis platforms, selecting biotin monomers based on modification position to ensure stable conjugation between biotin and oligonucleotides.
Monitor linker incorporation efficiency during synthesis to prevent steric hindrance from impairing biotin-streptavidin binding activity, especially for long-chain or complex modification combinations (e.g., concurrent fluorescent labeling).
-
Rigorous Quality Control
- Purity Validation: Confirm biotin modification accuracy via HPLC and mass spectrometry (MS), ensuring ≥99% purity. Separate unmodified or truncated sequences to guarantee batch consistency.
- Functional Testing: Include biotin-streptavidin binding efficiency assays, hybridization activity verification, and application compatibility tests.
-
Turnaround
Research-grade batches typically take 6-8 weeks, with expedited options (4-5 weeks) available. Scalable production from milligram to gram scales supports needs from laboratory research to preclinical trials.
-
Deliverables
Lyophilized biotin-modified oligonucleotides, accompanied by detailed quality reports, stability analyses, and customized usage guidelines.
Request a Customized Project Timeline
What We Can Offer?
Precise Custom Modification Solutions
Our expert team tailors biotin modification positions and linker arms to your experimental goals, such as intracellular tracking or targeted delivery, ensuring the oligonucleotide's original function remains intact and adapts to unique research needs.
High-Purity Synthesis Assurance
Leveraging advanced automated synthesis platforms, we strictly control reaction conditions to ensure ≥99% purity of biotin-modified oligonucleotides. Validated by HPLC and MS, batch-to-batch consistency exceeds 98%, guaranteeing reliable experimental data.
Comprehensive Functional Validation
Beyond testing biotin-streptavidin binding efficiency, we verify downstream critical applications like hybridization activity and cellular uptake efficiency, ensuring the modified oligonucleotides perform excellently in real experimental scenarios.
Flexible Production Scaling
Whether in milligram-scale early research or gram-scale preclinical development, we enable seamless transitions without redeveloping processes, meeting volume demands at every stage for biotin-modified oligonucleotides.
End-to-End Technical Support
From initial design consultations to troubleshooting during experiments, our professional team provides ongoing support. Drawing on extensive experience, we optimize protocols to facilitate smooth progress of your research projects.
[Legare Our Cutting-Edge Oligonucleotide Platform – Schedule a Consultation]
Customer Reviews

FAQs
Q1: What types of biotin modifications do you offer, and how do I choose the right one for my project?
A: We provide standard biotin (C6 spacer), biotin dT (internal), biotin-TEG (15-atom spacer), dual biotin, PC biotin (photocleavable), desthiobiotin-TEG, and biotin azide. Selection depends on your needs: biotin-TEG avoids steric hindrance for bead attachment; dual biotin suits high-sensitivity assays.
Q2: Does the position of biotin modification (5'-end, 3'-end, or internal) affect oligonucleotide function?
A: We design modifications to minimize interference. 5'/3'-end modifications (e.g., biotin-TEG) rarely impact binding, while internal biotin dT is placed in non-critical sequences. We validate with Tm profiling to ensure target affinity.
Q3: How does biotin-TEG differ from standard biotin in performance?
A: Biotin-TEG's 15-atom TEG spacer reduces steric hindrance, enhancing streptavidin binding, ideal for attaching to nanospheres or magnetic beads. Standard biotin (C6 spacer) works for general applications.
[Contact Our Team for More Information and to Discuss Your Project]
Reference
- Martín-Serrano, Ángela, et al. "Biotin-Labelled Clavulanic Acid to Identify Proteins Target for Haptenation in Serum: Implications in Allergy Studies." Frontiers in Pharmacology 11 (2020): 594755. DOI: 10.3389/fphar.2020.594755. Distributed under Open Access license CC BY 4.0, take partial screenshots of the picture.
