Cytogenetic Techniques
Cytogenetics is the science that studies the genetic material within the cell nucleus, namely the structure, function, and variation of chromosomes. Cytogenetic techniques are various methods used to detect and analyze chromosomes, including classical cytogenetic techniques and molecular cytogenetic techniques. Cytogenetic techniques have important application value in human genetic disease diagnosis and gene therapy, as they can help reveal the relationship between chromosomal abnormalities and gene variations and diseases, as well as provide individualized and precise treatment options. Notably, cytogenetic techniques still need to be constantly developed and innovated to improve their sensitivity, specificity, accuracy, and efficiency, and to combine with other genetic techniques to reveal more chromosomal variation information and biological significance.
Classical Cytogenetic Techniques
Classical cytogenetic techniques are methods that use conventional microscopy to observe and analyze the structure, number, and morphology of chromosomes. These techniques include chromosome preparation, chromosome analysis, chromosome banding, and karyotype analysis, which can produce characteristic patterns of light and dark bands along the chromosomes and help identify chromosomal abnormalities and genetic diseases.
Classical cytogenetic techniques have been widely used in the diagnosis of chromosomal abnormalities and genetic diseases for more than 50 years. They can provide valuable information on the gross structure and number of chromosomes in a cell, as well as the origin and inheritance of chromosomal aberrations. However, these techniques also have some limitations, such as low resolution, dependence on cell culture, difficulty in detecting submicroscopic changes and mosaicisms, and inability to identify gene mutations. Therefore, molecular cytogenetic techniques have been developed to overcome these drawbacks and provide more accurate and comprehensive information on the genome.
Molecular Cytogenetic Techniques
Molecular cytogenetic techniques are methods that use molecular probes to detect and analyze the genomic sequences and structure of chromosomes. These techniques include fluorescence in situ hybridization (FISH), comparative genomic hybridization (CGH), microarray technology, and gene chip technology, which can produce fluorescent signals or hybridization patterns on chromosomes or nuclei and help identify submicroscopic chromosomal abnormalities and gene variations. For example, FISH is a technique that uses fluorescently labeled DNA or RNA probes to hybridize with specific target sequences on chromosomes or nuclei. FISH can be used to detect chromosomal abnormalities, such as deletions, duplications, translocations, inversions, and aneuploidies, as well as gene expression and localization. FISH can be performed on metaphase chromosomes or interphase nuclei and can be combined with other techniques, such as chromosome banding, immunofluorescence, and spectral karyotyping.
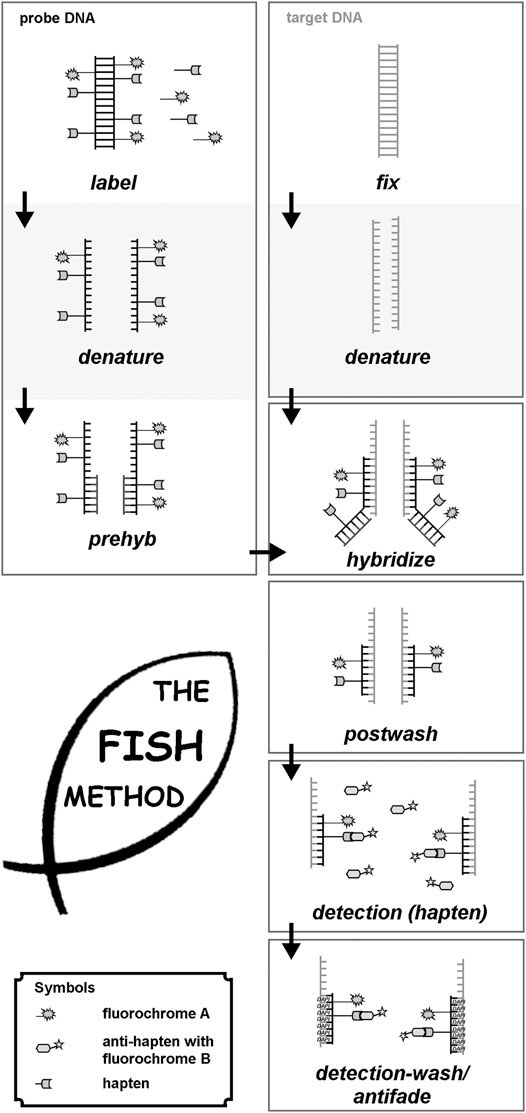 Fig.1 Principle of FISH. (Liehr T, 2021)
Fig.1 Principle of FISH. (Liehr T, 2021)
Molecular cytogenetic techniques have been developed to overcome the limitations of classical cytogenetic techniques and provide more accurate and comprehensive information on the genome. They can be used to detect copy number variations (CNVs), gene expression, methylation, and other genomic features in a genome-wide, high-throughput, and parallel manner. They can also be used to study the molecular mechanisms and pathways of chromosomal instability and genomic evolution.
Conclusion
Cytogenetic techniques are various methods used to detect and analyze chromosomes. These techniques include classical cytogenetic techniques and molecular cytogenetic techniques. These techniques have important application value in human genetic disease diagnosis and gene therapy, but they also face some challenges and future research directions. Classical cytogenetic techniques can provide information on the gross structure and number of chromosomes but have low resolution and difficulty in detecting submicroscopic changes and gene mutations. Molecular cytogenetic techniques can detect submicroscopic chromosomal abnormalities and gene variations, as well as other genomic features, but require specialized equipment and personnel and may be affected by DNA quality and hybridization conditions. How to improve the sensitivity, specificity, repeatability, and standardization of cytogenetic techniques; how to integrate different sources and levels of genomic data; how to explain the complex association between genomic variations and phenotypes; how to use cytogenetic techniques for individualized and precise treatment, etc., are all worthy of further discussion.
References
- Liehr T. Molecular Cytogenetics in the Era of Chromosomics and Cytogenomic Approaches. Front Genet. 2021 Oct 13;12:720507.
