Retrospective lineage tracing is particularly powerful for studying tumors because it leverages naturally occurring genetic and epigenetic alterations as intrinsic, permanent lineage markers within the tumor cells themselves. As cancer cells divide, they accumulate somatic mutations and epigenetic modifications, which are then inherited by their progeny. These alterations act as unique barcodes, allowing us to reconstruct the evolutionary history and clonal relationships within a tumor, even from archived or clinical biopsy samples where de novo labeling is impossible.
The core principle involves:
Mining Endogenous Tumor Barcodes: We identify and analyze the spectrum of naturally occurring genetic and epigenetic changes within tumor cells. For cancer research, key endogenous barcodes include:
- Somatic Mutations (SNPs, Indels): These random mutations accumulate over cell divisions and are powerful markers for tracking clonal expansion and divergence within a tumor. By analyzing shared and unique mutations, the phylogenetic relationships among tumor cells can be inferred.
- Copy Number Variants (CNVs): Gains or losses of large DNA segments are common in cancer and can represent significant clonal events, marking distinct tumor subclones.
- Mitochondrial DNA (mtDNA) Mutations: While less frequent, mtDNA mutations can also serve as stable clonal markers within tumor lineages.
- Epigenetic Modifications: Aberrant DNA methylation patterns are prevalent in cancer and can be stably inherited, providing additional layers of lineage information and potentially highlighting specific cancer-driving epigenetic events.
-
Tracing Clonal Expansion and Dissemination: By analyzing these inherited changes across different tumor regions, primary tumors, and metastatic sites, we can reconstruct the clonal architecture of a tumor. This allows us to:
- Identify the origins of tumor heterogeneity.
- Trace the routes and timing of metastatic dissemination.
- Pinpoint ancestral cancer-initiating cells or specific subclones responsible for tumor progression or drug resistance.
A critical advantage of our retrospective approach for in vivo tumor tracing is its non-invasive nature, as it requires no prior manipulation of the patient's cells. This enables the analysis of human tumor biopsies, allowing direct insights into patient-specific disease progression and therapeutic responses. Creative Biolabs' expertise in handling diverse clinical tumor samples, including FFPE tissues, ensures reliable and high-quality data generation. We employ proprietary methodologies to mitigatein vitro noise inherent in amplification processes, thereby maximizing the accuracy and resolution of lineage reconstruction from challenging tumor material.


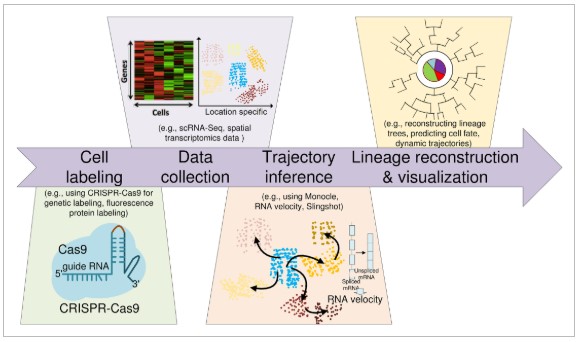 Fig.1 Computational cell lineage tracing workflow.1
Fig.1 Computational cell lineage tracing workflow.1


