T & B Cell based X-Ray Crystallography Assay Service
Cancer epitope analysis is the research that determines the specific cancer peptides that directly bind to cell receptors or antibodies and elicit immune responses. Creative Biolabs provides advanced and standard X-ray crystallography assays for the inspection of specific interactions between tumor antigen epitopes and immune cell receptors and antibodies.
Introduction What We Can Offer Workflow Why Creative Biolabs Customer Reviews FAQs Related Services Contact Us
Using X-ray Crystallography for Epitope Mapping
Protein antigen epitopes are classified into two kinds: linear and conformational epitopes. Linear epitopes consist of several continuous amino acid sequences, and conformational epitopes are not only amino acid sequences but also associated with the 3D structure of these peptides. In T cell immunity, the T cell receptor is responsible for the recognition of HLA-restricted antigen peptides, but most TCRs bind sub-optimally to tumor antigens. Moreover, most epitopes directly in contact with antibodies are complex, discontinuous conformational epitopes and thus should be screened by structural methods.
Discover how we can help - Request a consultation.
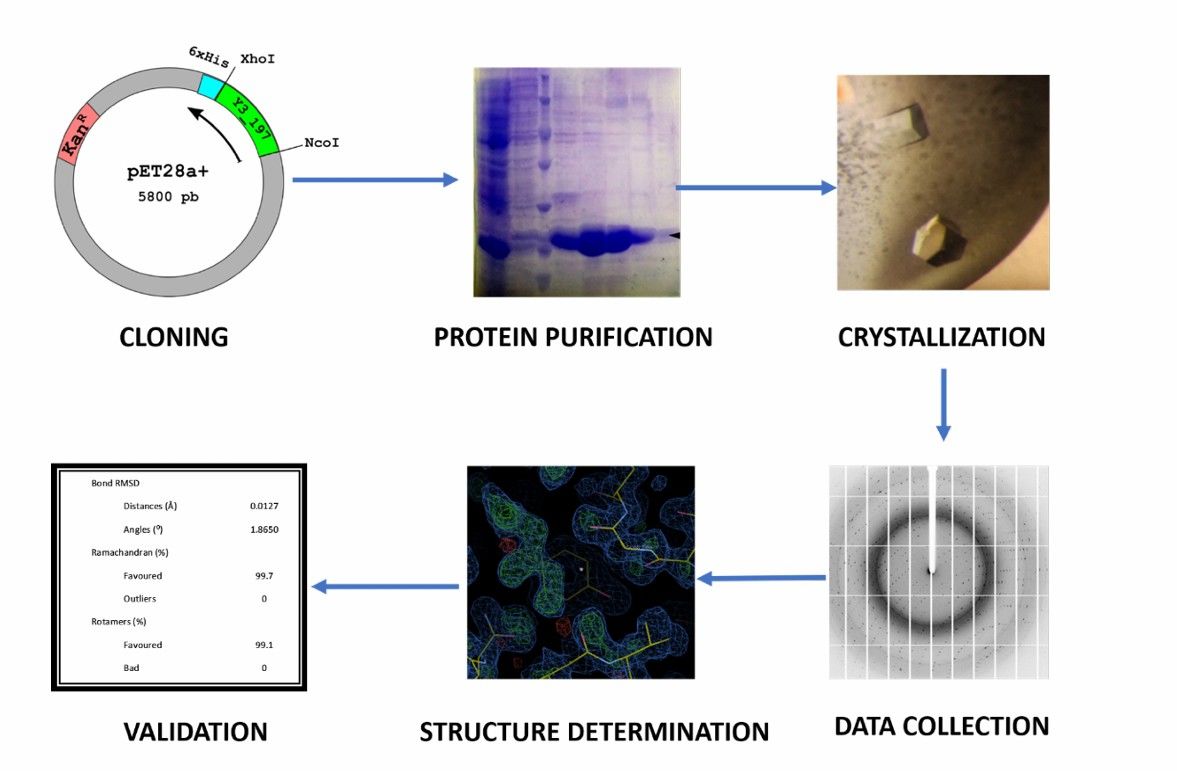 Fig.1 Main steps for protein structure determination using X-ray crystallography.1
Fig.1 Main steps for protein structure determination using X-ray crystallography.1
What We Can Offer
|
Key Features of X-ray Crystallography Assay
|
Antibody Format Can Be Used
|
-
Co-crystallization of the antibody-antigen complex.
-
Visualizing the antigen peptide-immune cell interactions in atomic resolution.
-
Facilitates the inspection of complex conformational epitopes.
-
Identifies the interaction domain of antigens against antibodies and receptors.
|
Any antibody format can be used for corresponding epitope analysis, including:
-
Fab
-
scFv
-
sdAb
-
Soluble TCR
-
Tailored Format
|
X-ray Crystallography Assay Services at Creative Biolabs
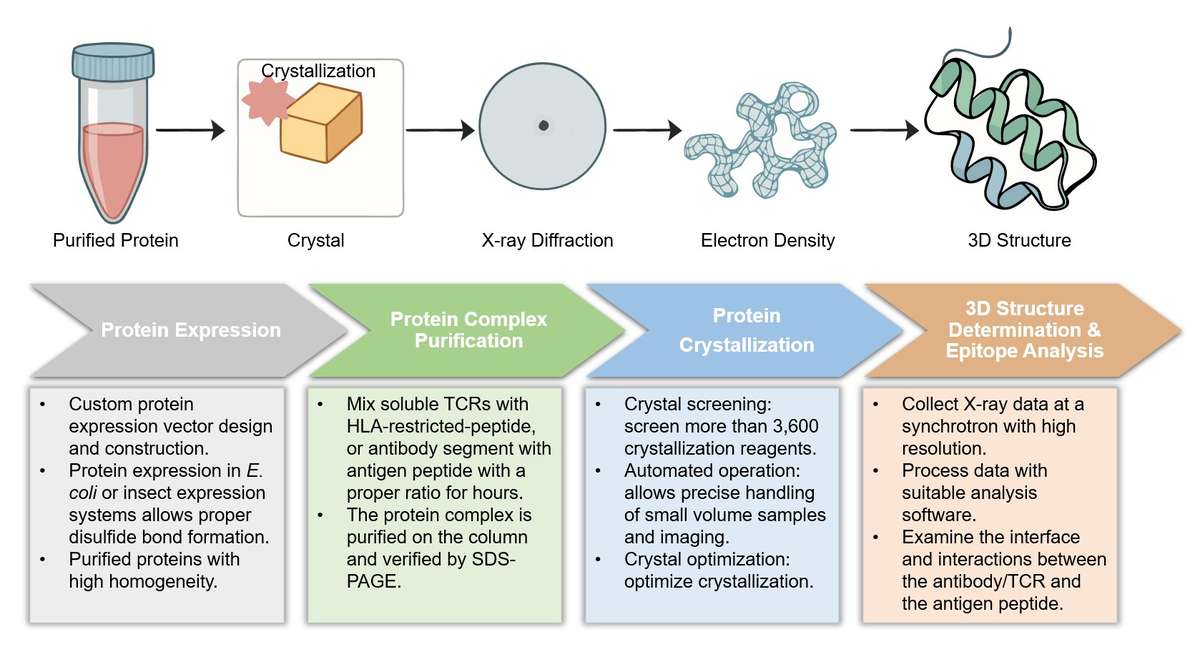
Highlights
Atomic-Level Visualization
Our assay provides high-resolution visualization of interactions between antigens and immune cell receptors. This enables the detailed inspection of complex conformational epitopes and the precise identification of an interaction domain.
Comprehensive Epitope Analysis
We facilitate the co-crystallization of antibody-antigen complexes and determine conformational epitopes for various receptors. This includes all antibody formats, such as Fabs, scFvs, and sdAbs, for comprehensive analysis.
Therapeutic Design Blueprint
The atomic-resolution data from our assay serve as a blueprint for rational drug design. This insight allows you to optimize lead molecules, enhance binding affinity, and accelerate your therapeutic development pipeline.
High-Quality Data
Our established platform and expertise ensure the production of high-quality, publication-ready data. This data is critical for supporting your research, patent filings, and regulatory submissions.
Discover how a partnership with us can streamline your research - Get a quote today.
Customer Reviews
-
Increased Precision
Using Creative Biolabs' X-ray crystallography service has significantly improved our epitope mapping for antibody-based therapies. The atomic-level detail clarified our MoA, a critical step we were unable to achieve with other methods. - Dr. Jhn A.
-
Tangible Results
Creative Biolabs' platform facilitated a breakthrough in our TCR development project. The high-resolution data on the TCR-peptide interaction enabled us to engineer a molecule with superior binding, which is now progressing through preclinical studies. - Dr. Rsa B.
FAQs
What if our protein is difficult to crystallize?
We have extensive experience with challenging proteins. Our high-throughput screening platform and expert optimization strategies are designed to overcome typical crystallization hurdles. We also offer tailored solutions for complex proteins, ensuring the best chance of success.
How do we get started, and what information do you need?
Getting started is simple! We'll need information about your target protein, your lead compound, and your project goals. We can also assist with protein production if you do not have a purified protein ready. To begin, simply reach out to our team to discuss your project requirements.
Related Services
TCR Identification
We can analyze the diversity and frequency of T-cell receptors to help identify and validate novel therapeutic targets and biomarkers for immunotherapy.
Learn More →
Immunogenicity Assessment of Candidate Neoantigens
We assess the ability of tumor-specific neoantigens to elicit a potent T-cell response, a critical step in developing personalized cancer vaccines and adoptive cell therapies.
Learn More →
How to Contact Us
Creative Biolabs possesses a leading technique in protein structure analysis and focuses on cancer epitope-receptor interactions using X-ray structure determination for years. Creative Biolabs works closely with our clients to provide tailored strategies for the structure and interaction determination, and provides detailed and accurate data and information on the complex. No more looking around, and contact us for a detailed quote.
Reference
-
Sanz-Gaitero, Marta, and Mark J van Raaij. "Crystallographic Structure Determination of Bacteriophage Endolysins." Current issues in molecular biology vol. 40 (2021): 165-188. Distributed under an Open Access license CC BY 4.0, without modification. https://doi.org/10.21775/cimb.040.165
For Research Use Only | Not For Clinical Use


 Fig.1 Main steps for protein structure determination using X-ray crystallography.1
Fig.1 Main steps for protein structure determination using X-ray crystallography.1


