Exosome Characterization
Creative Biolabs has extensive experience to offer a variety of exosome services including isolation, purification, and characterization. In order to better understand the characteristics of exosomes and to further facilitate the downstream therapeutic and diagnostic application, we offer various technologies applied for exosome characterization, including nanoparticle tracking analysis, FACS, western blotting, mass spectrometry (MS), and tunable resistive pulse sensing.
Exosome Characterization
Exosomes are a specific subclass of the extracellular vesicles that convey critical information in the form of small RNAs, mRNAs, bioactive lipids, and proteins. Acting as crucial mediators in cell-cell communication, exosomes play an important role in many aspects of physiology and disease. Their altered characteristics in many diseases, such as cancer, suggest their potential for diagnostic and therapeutic applications. Hence, isolation and characterization of exosomes from body fluids can provide very valuable information for early detection, disease monitoring and development of effective treatments against cancer and autoimmune diseases. In order to better define exosome characteristics and facilitate the downstream application, various technologies are applied for exosome characterization.
Exosome Characterization Services in Creative Biolabs
It is known that characterization of exosomes from body fluids can provide very valuable information for early detection, disease monitoring and development of effective treatments against cancer and autoimmune diseases. In order to better define exosome characteristics and facilitate the downstream application, Creative Biolabs has formulated a variety of protocols to better define the characteristics of exosome based on various technologies. The technologies applied for exosome characterization including nanoparticle tracking analysis, FACS sorting, western blotting, mass spectrometry (MS), as well as tunable resistive pulse sensing.
Nanoparticle Tracking Analysis-based Exosome Characterization
Nanoparticle tracking analysis is a highly sensitive method for visualization and analysis of exosomes. This method directly visualizes the light which is scattered from an exosome or microvesicle in liquid suspension. A finely focused laser beam passes through an optical interface such that it refracts through a thin layer of sample in liquid suspension. Light is scattered by the exosome/microvesicle in the path of the laser beam and is collected using a light microscope, and then imaged. Plots of scattered light spots and their speed of motion provide the data that facilitate the determination of total particle count and size distribution.
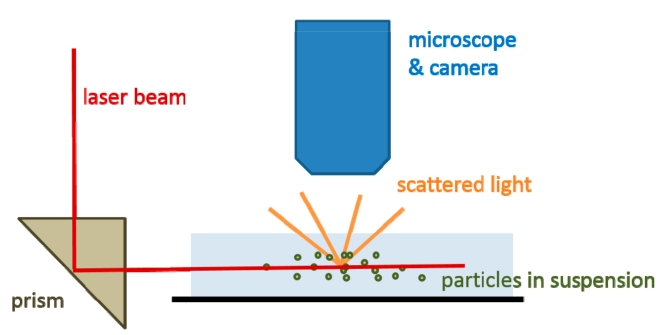 Fig. 1 A graphic representation of the nanoparticle tracking analysis (NTA) principle. (Szatanek, 2017)
Fig. 1 A graphic representation of the nanoparticle tracking analysis (NTA) principle. (Szatanek, 2017)
Exosomal Surface Markers-based Exosome Characterization
It is very essential to analyze the exosomal surface proteins, which may provide the critical information associated with the physiological states of their parental cell. Hence, standardization of exosome characterization is an essential step for reliable and reproducible results from assays and other downstream applications. Here, we demonstrate three methods for the analysis of exosomal surface marker.
-
Fluorescence-activated cell sorting (FACS)
FACS is the most common and prevalent tool used to analyze the different physical and chemical characteristics of exosome in suspension. This method utilizes light scattering properties of vesicles to analyze and sort individual exosomes using specific fluorescently labeled antibodies.
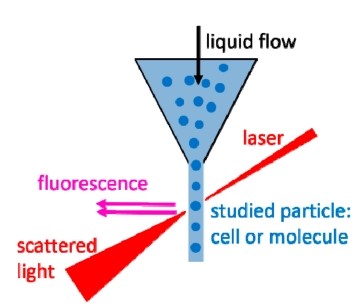 Fig. 2 The principal of flow cytometry. (Szatanek, 2017)
Fig. 2 The principal of flow cytometry. (Szatanek, 2017)
-
Western blotting
This method is a widely used analytical technique for detecting specific proteins in a sample of tissue homogenate or extract, as well as semi-quantitative estimation of protein derived from the size and color intensity of a protein band on the blot membrane. Thus, western blotting has been utilized to identify the presence and expression level of a specific single protein on the surface of exosomes.
-
Enzyme-linked immunosorbent assay (ELISA)
ELISA is a commonly used analytical biochemistry assay. It uses a solid-phase enzyme immunoassay (EIA) to detect the presence of a specific single protein on the surface of exosomes using a single or a pair of antibodies.

MS-based Exosome Characterization
MS is an analytical technique that ionizes chemical species and sorts the ions based on their mass-to-charge ratio. This method can be used to identify protein cargo in exosomes and determine whether the sample may contain protein aggregates or other contaminants.
Tunable Resistive Pulse Sensing (TRPS)-based Exosome Characterization
TRPS is a technique that allows high-throughput single particle measurements as colloids and/or biomolecular analytes driven through a size-tunable nanopore, one at a time. This method can be used to measure the size of exosome.
Features of Our Services
-
Advanced technology platforms
-
Accurate experimental data
-
Highly experienced professionals' team
-
Whole-process technical trace service
Based on the advanced technological platform, we can assist you to accurately analyze the characteristics of exosome derived from various sources. When you choose the exosome characterization of Creative Biolabs, you attain more than high-quality data delivery, you get in-depth scientific and technical support on everything from project planning to data interpretation from scientists with years of experience. If you have any demands in exosome characterization, please don’t hesitate to contact us for more information.
Reference:
-
Szatanek, R,; et al. The methods of choice for extracellular vesicles (EVs) characterization. International Journal of Molecular Sciences. 2017, 18(6):1153.
For Research Use Only. Cannot be used by patients.
Related Services:

 Fig. 1 A graphic representation of the nanoparticle tracking analysis (NTA) principle. (Szatanek, 2017)
Fig. 1 A graphic representation of the nanoparticle tracking analysis (NTA) principle. (Szatanek, 2017)
 Fig. 2 The principal of flow cytometry. (Szatanek, 2017)
Fig. 2 The principal of flow cytometry. (Szatanek, 2017)