Today, pancreatic cancer remains one of the most lethal malignancies facing the global population. The 5-year survival rate for patients with pancreatic ductal adenocarcinoma (PDAC) is only about 3%, and PDAC is difficult to treat surgically worldwide. In order to develop novel therapeutic strategies, researchers need to better understand the biology of PDAC at the molecular level. Recently, a paper was published in the international journal Cancer Science entitled “Identification of a novel target of SETD1A histone methyltransferase and the clinical significance in pancreatic cancer”, in which scientists from Tokyo Medical and Dental University and other institutions have identified a novel target gene that may have strong clinical significance in PDAC cases.
Previous research has shown that cancer cells express high levels of a specific enzyme, histone H3K4 methyltransferase, encoded by the SETD1A gene, which can add histones (essential structural components of chromatin) to specific target genes by methylating chemical groups. However, the molecular mechanism that triggers the overexpression of this enzyme and its effect on cancer cells is currently unknown.
When histones are methylated, target genes are activated, which is particularly relevant if SETD1A target genes support cancer development and progression. Therefore researchers are interested in understanding the key role that SETD1A overexpression plays in PDAC. “Although previous findings have shown that SETD1A is in overexpression mount in a variety of cancers, such as gastric and lung cancers, we do not know its specific molecular events in PDAC, and the targeting gene for SETD1A in PDAC has not yet been identified or recognized”, said researcher Takeshi Ishii.
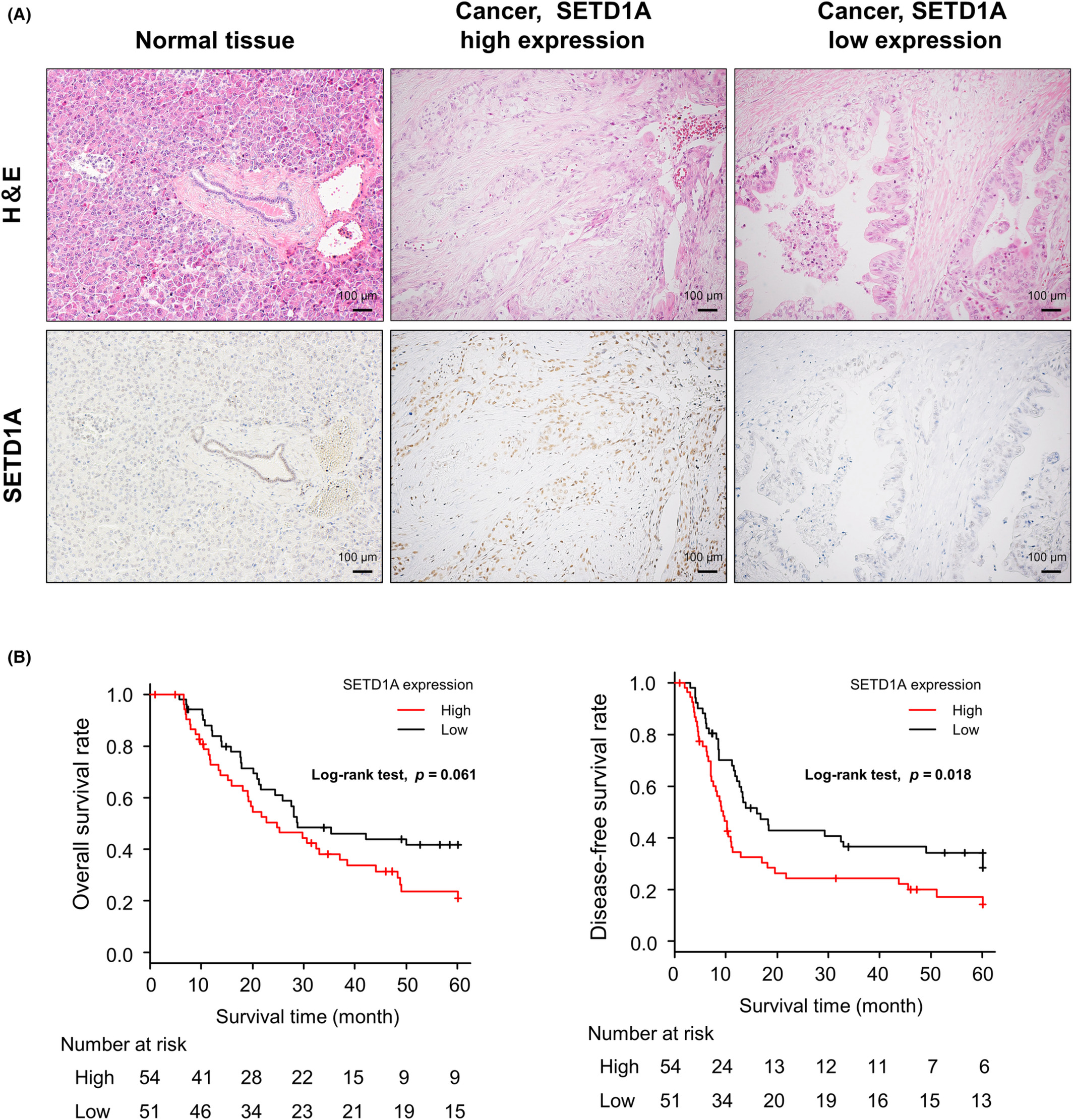
In this study, the researchers observed high levels of SETD1A expression in 51.4% of the human PDAC samples they analyzed, and they also determined that SETD1A may be an independent prognostic factor for disease-free survival, meaning that when the tumor is removed from the patient’s body, patients carrying high levels of SETD1A have a shorter duration of disease-free status compared with patients with low levels of SETD1A. The results of this study also demonstrate the clinical importance of SETD1A expression in PDAC.
The researchers then used artificially grown PDAC cells to analyze how altered SETD1A expression affected the behavior of the body’s cells, which had increased growth and migration capacity when overexpressed at SETD1A levels. In another group of PDAC cells, the researchers used molecular techniques to interfere with SETD1A expression and subsequently analyzed the other genes that were affected as a result. Using a special technique called RNA sequencing, the researchers analyzed the overall gene expression status after knocking down the expression of SETD1A and found that the expression level of another gene called RUVBL1 was also lower.
Further findings suggest that SETD1A methylates histones near the RUVBL1 gene and activates its gene expression, and that knocking down RUVBL1 expression in PDAC cells may have similar biological effects to those previously observed with interference with SETD1A. Analysis of survival rates showed that those carrying high levels of SETD1A and RUVBL1 PDAC patients had lower overall survival rates, suggesting that their co-expression is an important prognostic biomarker indicative of this cancer. Taken together, the findings of this paper provide a deeper understanding of the significance of SETD1A and RUVBL1 expression in PDAC, while also providing key details to assist clinicians in making critical clinical treatment decisions for patients experiencing severe diseases.
Reference
1. Ishii, Takeshi, et al. “Identification of a novel target of SETD1A histone methyltransferase and the clinical significance in pancreatic cancer.” Cancer Science 114.2 (2023): 463.
