NASH Target Development Service for PPAR Activators
Peroxisome proliferator-activated receptors (PPARs) are the ligand-activated transcription factors that are involved in the regulation of metabolism of lipid and lipoprotein metabolism as well as glucose homeostasis. It has been reported that PPARs are promising drug targets for NASH treatment. Creative Biolabs is very confident in offering the best services of target identification for drug discovery based on years of experience. Our services cover target screening, structural characterization, and functional profiling.
Introduction of PPARs
PPARs are a class of nuclear receptor proteins that are very essential ligand-activated transcription factors of nuclear hormone receptor superfamily and participate in the regulation of certain gene expression. The structure of PPARs is similar to steroid or thyroid hormone receptor and they are stimulated by small lipophilic ligands. It has been implicated that PPARs function as regulators of lipid, lipoprotein metabolism, and glucose homeostasis, influencing cellular proliferation, differentiation and apoptosis. PPARs mainly exist in three subtypes: α, γ and β/δ, which vary in their ligand specificity, tissue distribution, and physiological function. PPARα is highly expressed in liver, muscle, kidney, and heart, where it stimulates the beta-oxidative degradation of fatty acids. PPARγ is primarily expressed in intestine and adipose tissue and functions as an insulin sensitizer by regulating fatty acid uptake and lipid storage. PPARβ/δ is expressed more ubiquitously, but at relatively higher levels in the gastrointestinal tract, kidney, and skeletal muscle. The function of PPARβ/δ is mainly to facilitate the lipids decomposition and energy combustion.
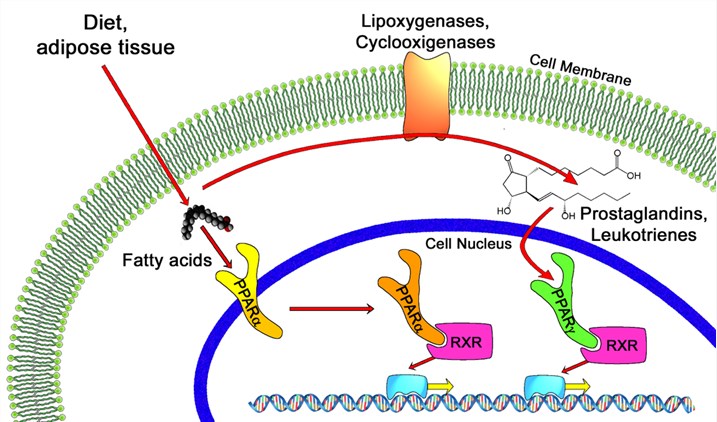 Fig.1 PPAR -alpha and -gamma pathways. Distributed under Open Access License CC BY 4.0, from Wiki, without modification.
Fig.1 PPAR -alpha and -gamma pathways. Distributed under Open Access License CC BY 4.0, from Wiki, without modification.
PPARs Activators Applied for NASH Treatment
Given the role of PPARs in glycometabolism and lipid metabolism, PPARs have become the promising targets for NASH treatment. More and more PPARs agonists have been exploited and evaluated in patients with NASH. Fibrates, the weak PPARα agonists, have hepatoprotective effects in rodent models of NASH. Furthermore, more potent PPARα agonists, including the selective PPARα modulator K-877 and the dual PPARα and PPARδ agonist GFT505 have been evaluated for NASH treatment. GFT505 also exhibits substantial hepatoprotective effects in rodent models of NASH. PPARδ agonists (such as MBX-8025) also display a good therapeutic effect in preclinical models of NASH by increasing hepatic lipid oxidation and insulin sensitivity as well as decreasing steatosis, inflammation and fibrogenesis. PPARγ agonists such as thiazolidinediones slow fibrosis progression in patients with NASH, but they have unwanted side effects such as weight gain, fluid retention, bone fractures, increased cardiovascular risk (for rosiglitazone) and increased bladder cancer risk (for pioglitazone) and so on. In order to decrease side effects, dual PPARα and PPARδ agonists combining the lipid-oxidizing and lipid-lowering properties of PPARα agonists and the insulin-sensitizing effects of PPARγ agonists have been developed for NASH treatment.
Creative Biolabs has a wealth of experience in drug target discovery. Equipped with advanced technologies and skilled expert team, we can offer high-quality target construction and custom target screening services to meet our clients’ demands precisely. Especially, our service items include antibody development (e.g. Phage Display & Antibody Library Services, Antibody Analysis Services, Antibody Engineering Services) and drug discovery. If you have any special needs in NASH services, please feel free to contact us for more details.
 For Research Use Only.
For Research Use Only.