Over the past two decades, protein-targeting degradation technologies (TPD), epitomized by PROTAC (proteolysis targeting chimera), have undergone rapid development and are considered one of the most promising breakthrough technologies for tackling difficult-to-drug targets. Traditional PROTAC technology leverages the catalytic action of E3 ubiquitin ligases to achieve specific degradation of targets via the ubiquitin-proteasome pathway. However, limited by the finite variety of available E3 ligases and the substrate specificity of the proteasome, recent years have seen the emergence of various lysosome-based TPD strategies. These include LYTAC technology, which targets cell membrane proteins or extracellular proteins, and strategies utilizing autophagy pathways for the degradation of solid aggregates or organelles, such as ATTEC (Autophagosome-Tethering Compound) and AUTOTAC. The advent of these technologies has further propelled the development of the TPD field and offered potential therapeutic strategies for a variety of serious diseases.
With the burgeoning field of “liquid-liquid phase separation,” an increasing number of studies have shown that key proteins in the degradation process can form “degradation condensates” through liquid-liquid phase separation. These condensates act as degradation factories, concentrating degradation-related functional proteins and substrates to be degraded in localized compartments, thereby efficiently executing degradation functions. The most representative of these degradation condensates is the formation of p62 bodies by autophagy receptor protein p62 and polyubiquitin chains. These structures provide anchor points for the attachment and extension of isolation membranes, initiating the subsequent autophagy process (Figure 1). Moreover, p62 also participates in ubiquitin-proteasome pathway-related degradation condensates, serving as a bridge between the proteasome and autophagy pathways. If it were possible to specifically recruit protein targets to these degradation condensates, it might bypass enzymatic catalytic steps (e.g., the catalytic reaction of E3 ubiquitin ligases) and achieve effective degradation by merely bringing them into close spatial proximity.
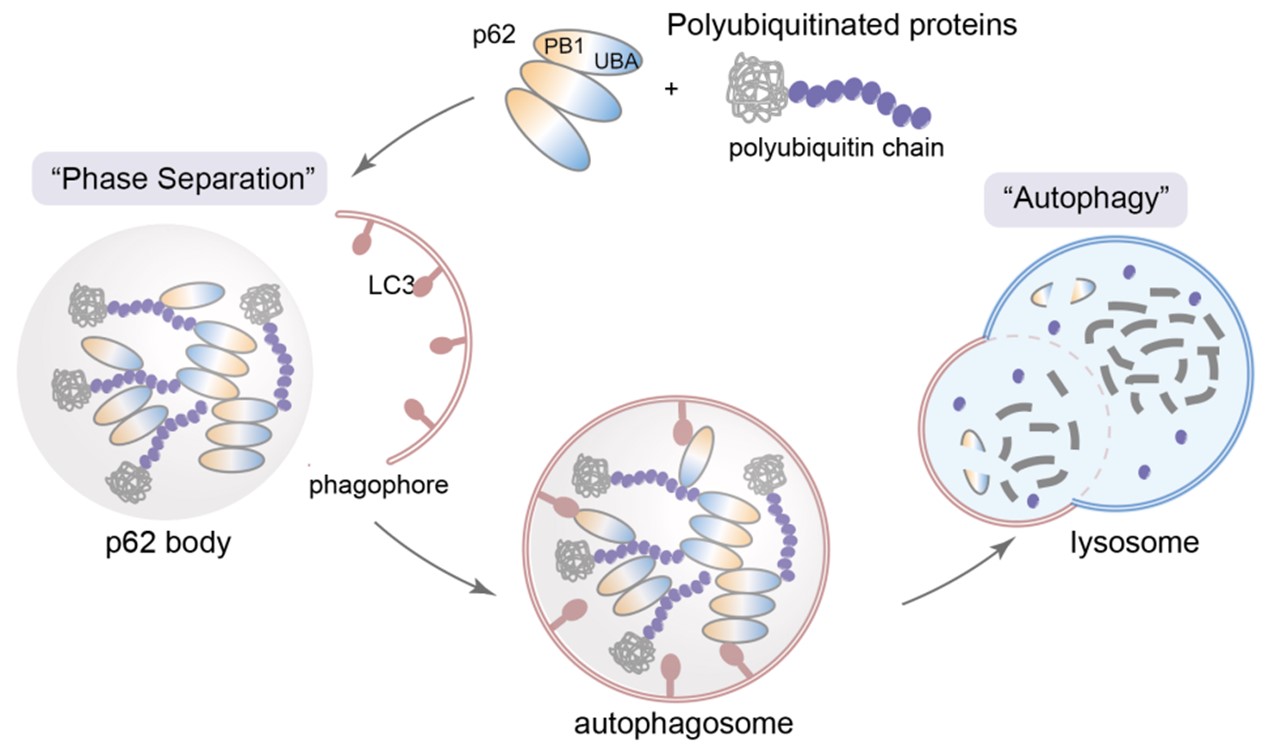
Figure 1. Autophagy degradation process mediated by p62 degradation condensate
Recently, the research group of Li Pilong and Li Zengpeng of the third Institute of Oceanography of the Ministry of Natural Resources published a cooperative research paper on an integrated targeted protein degradation platform in Cell Research. The paper reports a novel protein-targeting degradation technology named PDF-Bin. The core component of this technology is a bispecific antibody capable of simultaneously binding to the p62 protein and the target protein. It can specifically recruit the target protein to p62 degradation condensates, thereby achieving targeted degradation functionality (see Figure 2).
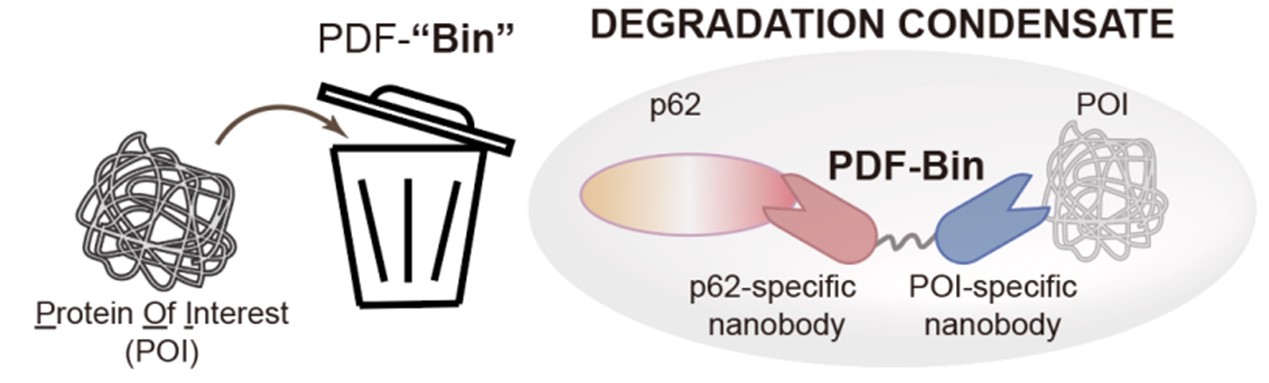
Figure 2. A schematic diagram of PDF-Bin
PDF-Bin is named after P62 degradation factory based on a bispecific antibody, comprising an antibody that specifically binds to p62 (Pn), an antibody that specifically binds to the target protein, and a flexible linker connecting them. To obtain p62-specific antibody, researchers utilized phage display technology combined with colocalization analysis through microscopy, resulting in the identification of three antibody specifically binding to p62 (A1E, D9A, and E12C). PDF-Bin constructs created by fusing these three Pns with an EGFP-tagged antibody Gn (A1E-Gn, D9A-Gn, and E12C-Gn) all demonstrated effective degradation of EGFP-TDP43 protein.
Subsequently, the researchers tested the degradation capability of A1E-Gn against a series of targets fused with the EGFP tag. The results showed that A1E-Gn could degrade target proteins located in the nucleus, cytoplasm, cell membrane, and those forming solid aggregates, proving that PDF-Bin has a broad range of applications.
To further explore the properties of PDF-Bin, the researchers constructed respective PDF-Bin fusion proteins targeting EGFP-TDP43 and the endogenous STAT3. Surprisingly, the degradation of EGFP-TDP43 by PDF-Bin depended on the proteasome pathway, while the degradation of STAT3 was mediated through the autophagy-lysosome pathway. This result suggests that PDF-Bin selects the most suitable degradation pathway based on the inherent characteristics of the target protein, thus achieving more efficient degradation (see Figure 3).
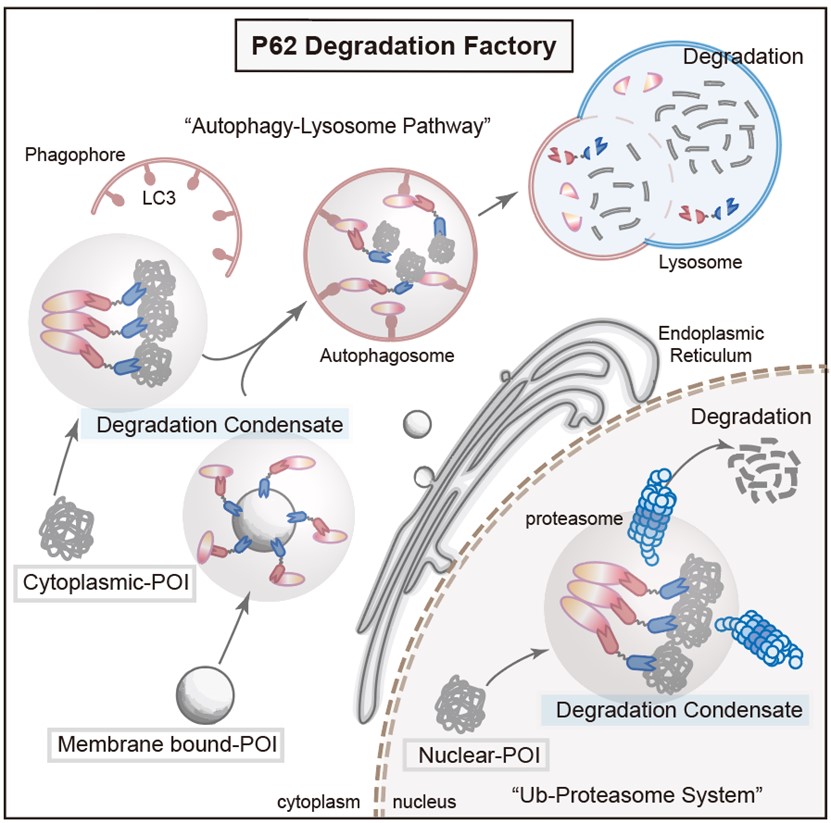
Figure 3. A schematic diagram of PDF-Bin degradation mechanism
Lastly, the researchers preliminarily explored the therapeutic potential of PDF-Bin. STAT3, a classic target in cancer therapy, is widely activated in various cancers and regulates the proliferation and differentiation of cancer cells. By specifically degrading STAT3 protein in non-small cell lung cancer cell lines using PDF-Bin, tumor cell proliferation and migration were effectively inhibited, offering new strategies for the development of novel anti-tumor drugs.
In summary, this study developed a protein-targeting degradation strategy based on bispecific antibodies. It is effective in degrading target proteins across various subcellular localizations and different mobilities. Additionally, it integrates the ubiquitin-proteasome and autophagy-lysosome pathways, autonomously selecting the optimal degradation pathway based on the characteristics of the target protein.
Reference:
Jia, Wen, et al. “An all-in-one targeted protein degradation platform guided by degradation condensates-bridging bi-specific nanobodies.” Cell Research (2024): 1-4.
