Protein degradation is an important mechanism by which organisms regulate the amount and activity of proteins in cells. The ubiquitin-proteasome system is involved in regulating important physiological or pathological processes such as cell growth, proliferation, migration, differentiation, and death. The 2004 Nobel Prize in Chemistry was awarded to three Israeli scientists for their early discovery of the mechanism of ubiquitination-enzymatic intracellular protein degradation. Targeted protein degradation (TPD) is a novel drug development strategy that uses specifically recognized small molecules or macromolecules to form a targeting chimera (PROTAC) bifunctional molecule with a ubiquitin-proteasome. It can “hijack” the degradation process of cells and degrade key target proteins from cells to control key protein molecules in cells and explore their efficacy. Currently, large-scale screening is generally used to obtain chemical small molecule-mediated methods that specifically bind to target degradation proteins. However, its limitation lies in the need to find high-affinity small molecule ligands for the target protein, which is very difficult for some “untargetable” proteins. Scientists have achieved new results by using single-domain antibody targeted degradation technology to explore different subcellular functions of intracellular proteins. Relevant research results were recently published in the journal.
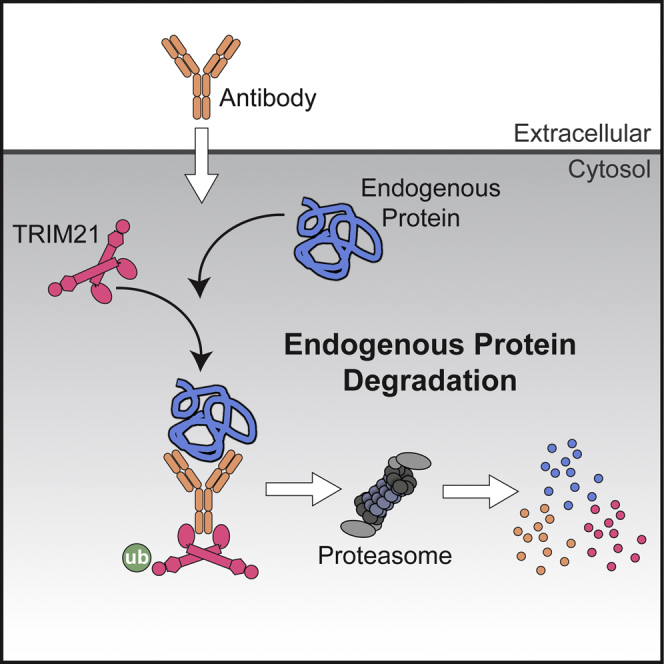
Fig. 1 Endogenous proteins degradation. (Dean Clift, 2017)
Based on the previous construction of a camel-derived single domain antibody library and high-throughput screening, this study created a single domain antibody-based molecular toolkit-ubiquitin proteasome system – Nb4A-Fc-T2a-TRIM21. This system can target and degrade Survivin protein located in the cytoplasm and nucleus through the ubiquitination pathway, thereby exploring the potential functions of Survivin protein in different subcellular localizations. This reveals the different functions and activities in its different subcellular structures and provides a scientific basis for the further development of drugs that precisely target its subcellular structures.
Most of the systems reported so far use chemical molecules to regulate protein degradation, with poor temporal and spatial resolution. Using antibodies instead of small molecule targeting will result in a larger molecular weight and poor membrane permeability, resulting in poor bioavailability and poor druggability. There are issues with the precision of regulation and possible off-target toxicity, as well as efficient cell production and cost when using whole antibodies. Single domain antibodies (sdAbs) have fully functional antigen-binding fragments that can target target proteins just like antibodies. They have the advantages of a small molecular weight, easy expression, high stability, and strong specificity. The Nb4A-Fc-TRIM21 system created in this study can rely on ubiquitination to target and degrade the Survivin protein located in the cytoplasm and nucleus and has the advantage of being fast and effective. Experimental results show that Survivin in the cytoplasm mainly exerts anti-apoptotic effects by directly or indirectly inhibiting the caspase pathway, while Survivin in the nucleus mainly promotes cell proliferation and participates in cell cycle regulation. This lays the theoretical foundation for an in-depth study of the mechanism of Survivin protein and the discovery of feasible and effective tumor drug targets. At the same time, this study also demonstrates the potential for wider application of single domain antibodies in basic research. (Learn more about our One-Stop Solution for Anti-Membrane Protein sdAb Development)
Reference
Wang, S., Xu, Y., Chan, H. F., Kim, H. W., Wang, Y., Leong, K. W., et al. (2016). Nanoparticle-mediated inhibition of survivin to overcome drug resistance in cancer therapy. J. Control. Release 240, 454–464.
Fan, L., Sun, G., Ma, T., Zhong, F., and Wei, W. (2013). Melatonin overcomes apoptosis resistance in human hepatocellular carcinoma by targeting survivin and XIAP. J. Pineal. Res. 55, 174–183.
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096.
Clift, D., McEwan, W. A., Labzin, L. I., Konieczny, V., Mogessie, B., Jameset, L. C., et al. (2017). A method for the acute and rapid degradation of endogenous proteins. Cell 171, 1692–1706.e18.
Miao H, Liu C, Ouyang H, Zhang P, Liu Y, Zhang C, Deng C, Fu Y, Niu J, Zheng W, You F, Yang Y, Ma X. A nanobody-based molecular toolkit for ubiquitin-proteasome system explores the main role of survivin subcellular localization. Front Bioeng Biotechnol. 2023 Jan 20;10:952237. doi: 10.3389/fbioe.2022.952237. PMID: 36743654; PMCID: PMC9895104.
