Whole-genome doubling (WGD) is a recurring event in human cancer that promotes chromosomal instability and the acquisition of aneuploidy, but the three-dimensional (3D) assembly of chromatin in WGD cells and its contribution to the oncogenic phenotype remains unknown. In a recent study published in the journal Nature, entitled “Whole-genome doubling drives oncogenic loss of chromatin segregation”, scientists from the Swiss Federal Institute of Technology in Lausanne and other institutions have found that WGD cells are more likely to have chromatin doubling. Scientists have uncovered new clues that WGD drives cancer discovery and that WGD can also affect the 3D architecture of chromatin in cells through a phenomenon called loss of chromatin segregation (LSC).
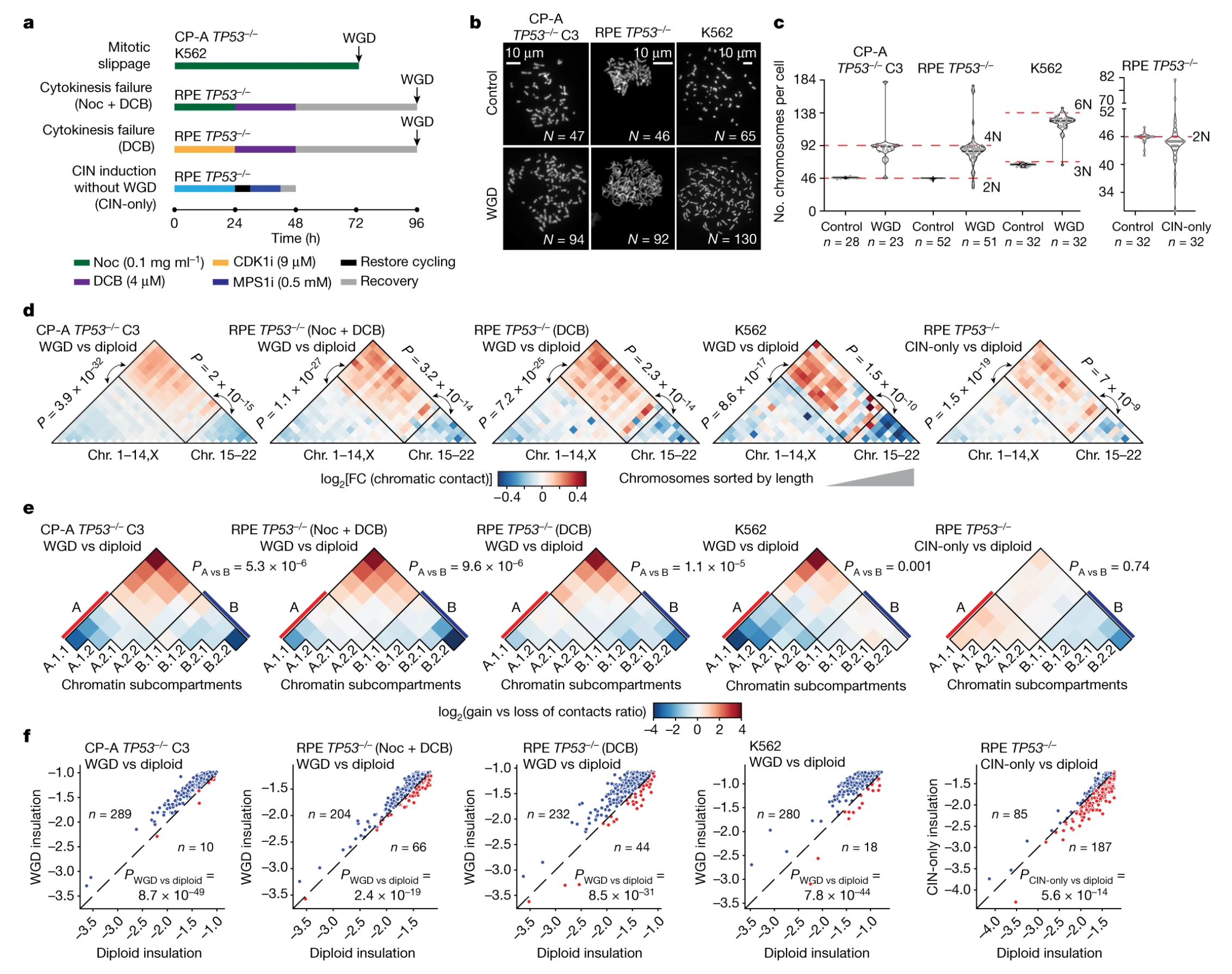
Fig. 1 Whole-genome doubling drives oncogenic loss of chromatin segregation (Lambuta, 2023)
A cell contains 2-3 meters of DNA, which means that the only way to store it is to pack it into tightly packed coils, and the solution is chromatin, a special protein complex wrapped in DNA called histones. In 3D space, this complex can gradually fold into a multi-layered organization of loops, structural domains, and compartments, which make up what we know as chromosomes. The organization of chromatin is closely linked to both gene expression and the proper functioning of the cell, so any problems with the structure of chromatin can lead to disadvantages, such as the development of cancer.
A common event that occurs in approximately 30% of all human cancers is WGD, where entire sets of chromosomes are duplicated in a cell. WGD causes genomic instability in the cell, leading to chromosomal alterations and other mutations that contribute to the development of human cancers.
In the paper, the researchers investigated cells lacking the p53 gene, the tumor suppressor gene, which predisposes them to WGD. They found that WGD leads to reduced levels of segregation of chromatin structural elements, such as loops, structural domains, and compartments, as well as disrupting their careful assembly in the cell. These changes result that mixtures of genetic material that normally remain separate change the position of genomic regions in 3D space, in what has been termed sub-compartment repositioning, which may create the conditions for the activation of oncogenes.
The researchers also found that the effect of WGD on chromatin assembly is largely dependent on chromosomal alterations, implying that chromatin segregation and chromosomal instability can act as complementary mechanisms that work together to promote cancer development.
In summary, the results of this paper suggest that LSC may initiate chromatin conformational changes that ultimately lead to oncogenic epigenetic and transcriptional modifications, suggesting that chromatin evolution may be a major marker of WGD-driven cancers. This study provides a new way for researchers to observe the role of WGD and chromatin assembly in carcinogenesis, and in the future, highly multiplexed single-cell molecular mapping combined with barcoding techniques and novel computer tools may help unravel the key role that disruption of chromatin 3D structure plays in transforming cells into cancerous ones.
Reference
1. Lambuta, Ruxandra A., et al. “Whole-genome doubling drives oncogenic loss of chromatin segregation.” Nature 615.7954 (2023): 925-933.
