Vesiclepedia (http://www.microvesicles.org) stands as a comprehensive, web-based database, offering a compilation of DNA, RNA, proteins, lipids, and metabolites that have been identified or associated with extracellular vesicles (EVs) and extracellular particles (EPs). Both EVs and EPs encompass proteins, nucleic acids, lipids, and metabolites, playing pivotal roles in numerous biological functions. Vesiclepedia meticulously catalogs information from both published and unpublished research. Recently, researchers from La Trobe University in Australia have diligently updated the contents of the Vesiclepedia database. Presently, the database provides quantitative data for 62,822 entries across 47 EV studies. Users can access the dataset through a tab-delimited file or a database utilizing the FunRich’s Vesiclepedia plug-in. The pertinent details were published online on November 11 in Nucleic Acids Research, an international journal specializing in biochemistry and molecular biology, under the titled “Spatial exosome analysis using cellulose nanofiber sheets reveals the location heterogeneity of extracellular vesicles.”

Extracellular vesicles (EVs), nanometer-sized membrane vesicles released into the extracellular space by cells, are known to be widely secreted across all domains of life, from archaea to eukaryotes. Additionally, some mammalian cells reportedly release extracellular particles (EPs). Over the past two decades, EVs have gained recognition for their pivotal roles in multiple pathophysiological processes, owing to their signaling capabilities. Notably, these signaling molecules not only mediate communication between distant cells within individuals but also play intricate roles in inter-species and inter-kingdom communication. EVs are classified into diverse subtypes based on their occurrence, origin, size, cargo composition, and functionality. Exosomes (30–150 nm) originate from the fusion of multivesicular endosomes with the plasma membrane, while ectosomes (100–1000 nm) are formed through protrusions from the plasma membrane. Exosomes and extracellular bodies are actively secreted by living cells, whereas apoptotic bodies (50–5000 nm) are released by long-standing apoptotic or dying cells. Similar to exosomes, apoptotic bodies also originate due to protrusion from the plasma membrane. Migrasomes (500–3000 nm) are released by migrating cells, and large oncosomes (1000–10 000 nm) are released by deforming tumor cells. In EPs, including exomeres and supermeres smaller than 50 nm, the mechanism remains unknown.
 Figure: Subtypes of EVs are categorized based on their generation pathways. Exosomes, ranging from 30 to 150 nm, originate from the endosomal pathway. Microvesicles/exosomes, ranging from 100 to 1000 nm, form through extrusions from the cytoplasmic membrane. Migrant bodies, vesicles sized from 500 to 3000 nm, are left behind by migrating cells. Apoptotic bodies with diameters up to 1–5 μm, constitute the largest category of EV subtypes and form through swelling and protrusion of the apoptotic membrane during cell death. Large tumor bodies, ranging from 1 to 10 μm, are sizable vesicles derived from cancer cells. Broadly classified by operational terminology and size, EVs and particles are categorized into large EVs, small EVs, and EPs.
Figure: Subtypes of EVs are categorized based on their generation pathways. Exosomes, ranging from 30 to 150 nm, originate from the endosomal pathway. Microvesicles/exosomes, ranging from 100 to 1000 nm, form through extrusions from the cytoplasmic membrane. Migrant bodies, vesicles sized from 500 to 3000 nm, are left behind by migrating cells. Apoptotic bodies with diameters up to 1–5 μm, constitute the largest category of EV subtypes and form through swelling and protrusion of the apoptotic membrane during cell death. Large tumor bodies, ranging from 1 to 10 μm, are sizable vesicles derived from cancer cells. Broadly classified by operational terminology and size, EVs and particles are categorized into large EVs, small EVs, and EPs.
In terms of molecular composition, EVs harbor a diverse array of proteins, lipids, and nucleic acids, facilitating their secure transport from donor to recipient cells. As the functionality of EVs is contingent on their contents, the cargo evolves with the changing pathophysiological conditions of the host cells from which they emanate. Thus, systematically cataloging EV cargo—encompassing nucleic acids, proteins, metabolites, and lipids—under various conditions proves instrumental in discerning EV fingerprints unique to specific tissues, cell types, or pathologies.
This article presents an update to the Vesiclepedia database (http://www.microvesicles.org), an online repository housing RNA, proteins, lipids, and metabolites identified in EVs and extracellular particles (EPs). Moreover, Vesiclepedia incorporates DNA data related to EVs and EPs from 52 studies, including entries for 167 oncogenes and mutated genes.
Vesiclepedia is a fusion of MySQL as the backend database, the Zope content management system, and Python as the programming language linking the database and content management system. The website empowers users to seamlessly query or browse the database and download the entire dataset as a free tab-delimited file. Whether querying a gene or exploring EV studies, users gain access to supplementary information such as gene summaries, EV isolation methods, flotation density (if available), EV subtype and sample source. Cargo data in Vesiclepedia result from community annotation or meticulous manual curation of docum. While databases like ExoCarta focus on exosomes or small EVs, such as ExoCarta, Vesiclepedia (http://www.microvesicles.org) spans cargos from various EV subtypes and EPs.
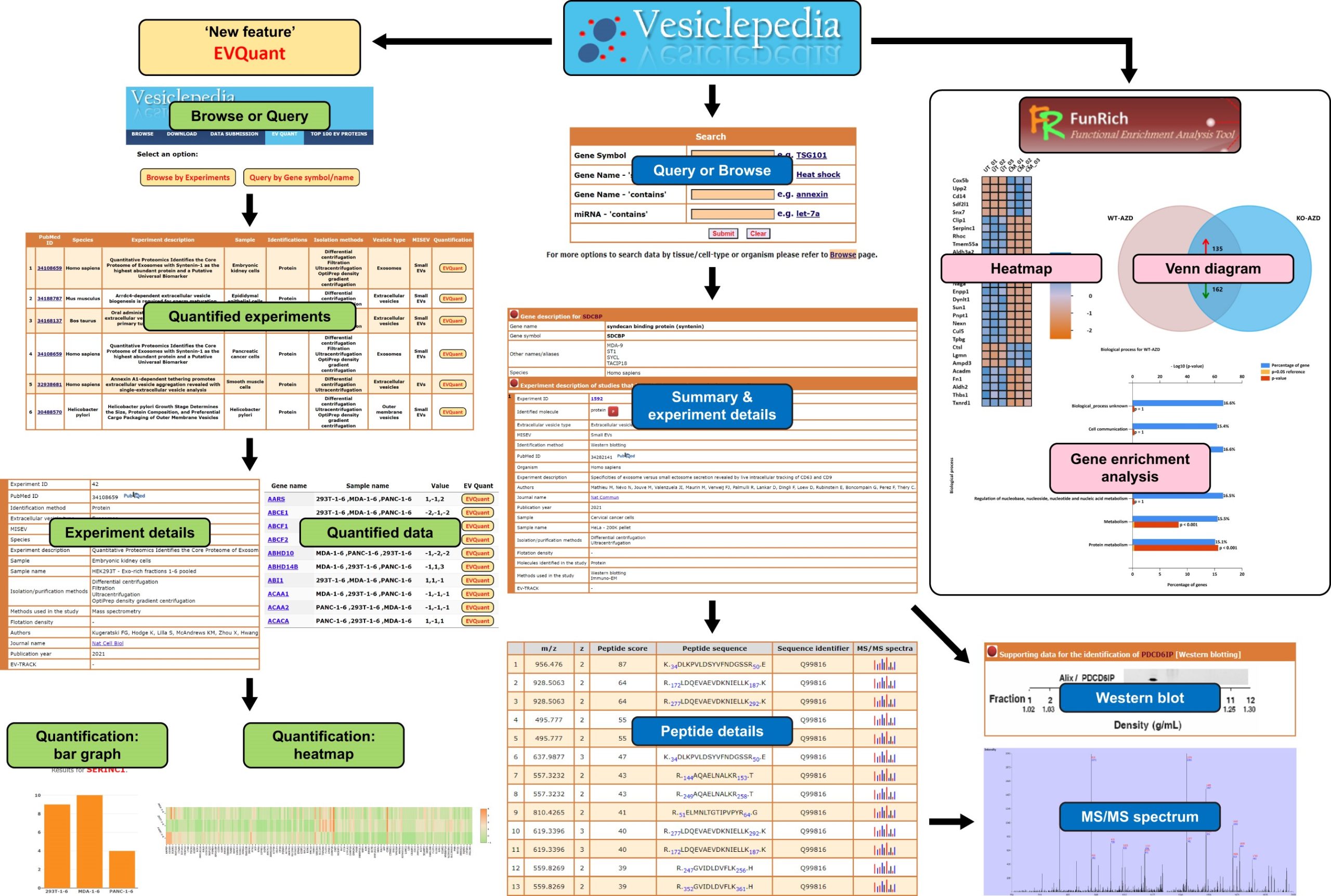 A snapshot of the Vesiclepedia page, featuring the EV-enriched protein TSG101, exemplifies the platform’s capabilities. A search for a molecule of interest provides a gene summary page, presenting data on the molecule, and experimental details from studies identifying it. This includes Western blotting data, mass spectrometry-based detailed peptide information, and tandem MS spectra where applicable. EV-QUANT, an integral part of Vesiclepedia, offers quantitative data for EV research, including data tables and visualizations like heat maps and bar graphs for enriched cargoes and gene quantitation. Researchers can utilize the Vesiclepedia plug-in in FunRich for automated data retrieval for downstream analysis.
A snapshot of the Vesiclepedia page, featuring the EV-enriched protein TSG101, exemplifies the platform’s capabilities. A search for a molecule of interest provides a gene summary page, presenting data on the molecule, and experimental details from studies identifying it. This includes Western blotting data, mass spectrometry-based detailed peptide information, and tandem MS spectra where applicable. EV-QUANT, an integral part of Vesiclepedia, offers quantitative data for EV research, including data tables and visualizations like heat maps and bar graphs for enriched cargoes and gene quantitation. Researchers can utilize the Vesiclepedia plug-in in FunRich for automated data retrieval for downstream analysis.
With this update, Vesiclepedia now contains 3,533 EV studies, more than doubling the count since the last update in 2019. The integration of data from 56,911 protein entries, 50,550 RNA entries, 3839 lipids, 192 metabolites, and 167 DNA entries further enriches the repository. Currently, Vesiclepedia provides quantitative data across 62,822 entries from 47 studies, cataloging 252 sample sources from 56 organisms.
Reference:
Chitti SV, Gummadi S, Kang T, et al. Vesiclepedia 2024: an extracellular vesicles and extracellular particles repository [published online ahead of print, 2023 Nov 11]. Nucleic Acids Res. 2023;gkad1007. doi:10.1093/nar/gkad1007
Related Services:
Exosome Cargo Loading Services
