Extracellular vesicles (EVs), actively secreted from cancer cells and host cells into the circulation, are emerging as one of the front-runners in diagnostics for early cancer detection, disease monitoring, and treatment evaluation. Researchers from the Center for Systems Biology at Massachusetts General Hospital published a review in the journal Trends in Molecular Medicine, outlining EVs and the basic principles for early cancer detection, studying emerging technologies for single EV analysis and their application in early cancer detection.
Does someone have cancer that will grow malignantly and eventually metastasize? This is a critical question that the medical community, including medicine, oncology, preventive medicine, medical epidemiology, and health care cost communities, have been grappling with. It is known that growing cancer releases a variety of components into the circulation, including whole cells (i.e., circulating tumor cells (CTCs)), tumor-derived DNA (ctDNA), various EVs, proteins, and metabolites. The premise of “liquid biopsy” is to use these components derived from tumor cells to make a diagnosis without relying on medical imaging or tumor biopsies. Currently, key questions in early cancer diagnosis are: Which blood component is best to detect? How common is a given marker (rare or ultra-rare)? And how specific is that marker for a given cancer type?
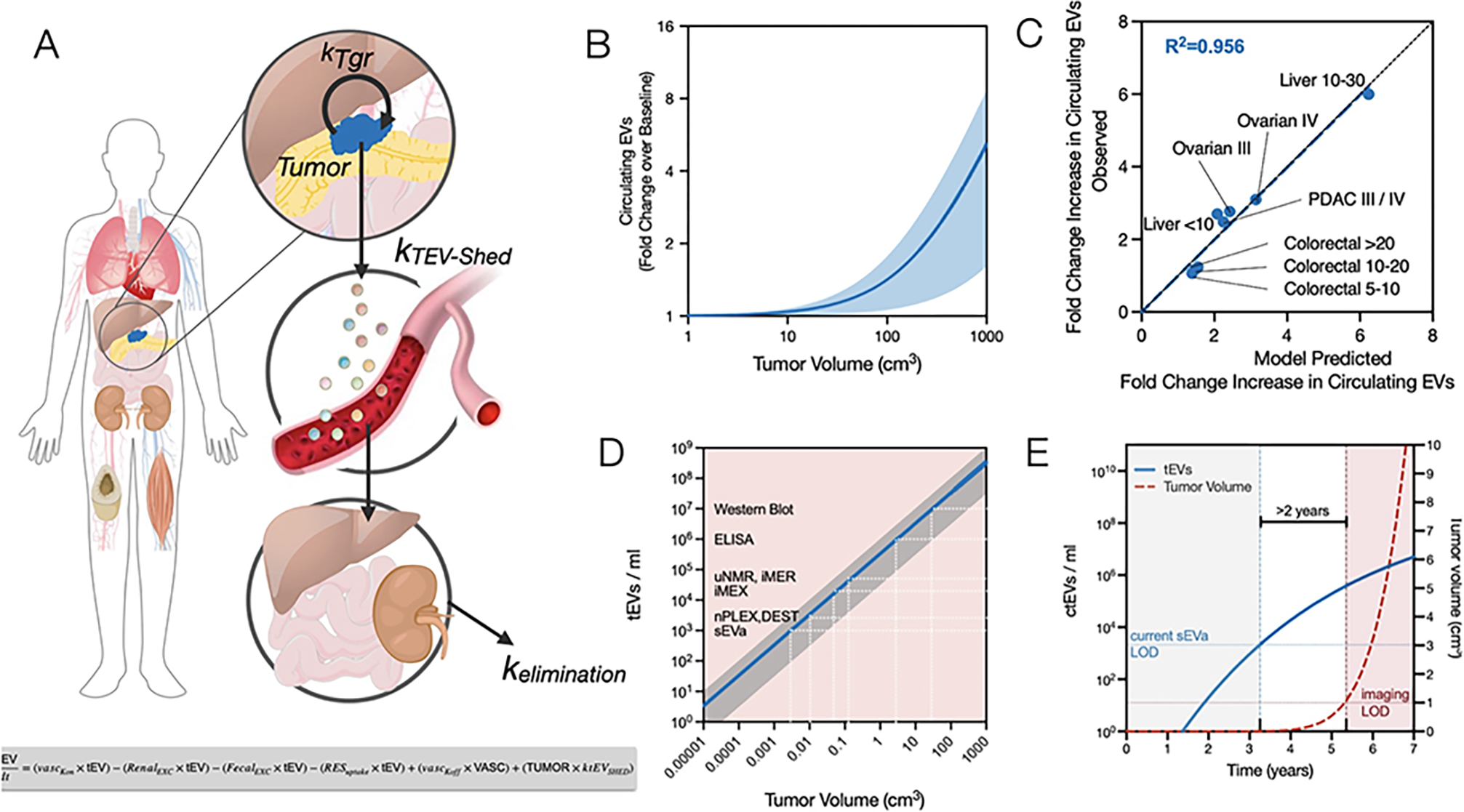
EVs have emerged as leaders in early cancer detection, disease monitoring, and treatment evaluation. One advantage of EVs is their continuous shedding by dividing tumor cells, with indications suggesting a correlation between higher tumor metabolic rates and increased EV production. Unlike ctDNA diagnostics, EV shedding occurs in living cells and is not rely on tumor cell death. Therefore, EV production likely serves as an ongoing event and represents one of the earliest sources of biomarkers in cancer cells. In advanced melanoma, the DNA of mutant alleles BRAFV600E and cKITD816V was found not to be associated with EVs. However, the authors believe this discrepancy may not hold true in patients with early-stage cancer. Direct comparisons between exosomal DNA and ctDNA in other studies have revealed that exosomal DNA is superior to ctDNA in detecting mutations in pancreatic and lung cancer. Furthermore, EVs address a significant issue that has hindered the wider use of RNA and proteins by protecting cargo from degradation in the bloodstream. Additionally, EVs can encapsulate multiple cargoes within vesicles, ultimately indicating the tissue of origin.
Recent studies have demonstrated that circulating tumor-derived EVs (ctEVs) are heterogeneous, with only a minority carrying tumor-specific biomarkers such as mutant proteins. Detecting of these highly specific but rare ctEVs among a large population of host cell-derived EVs may require single EV analysis (sEVA). This review highlights recent advances in sEVA, focusing on clinical applications, discussing current limitations, and proposing further work needed to enhance existing and emerging toolkits.
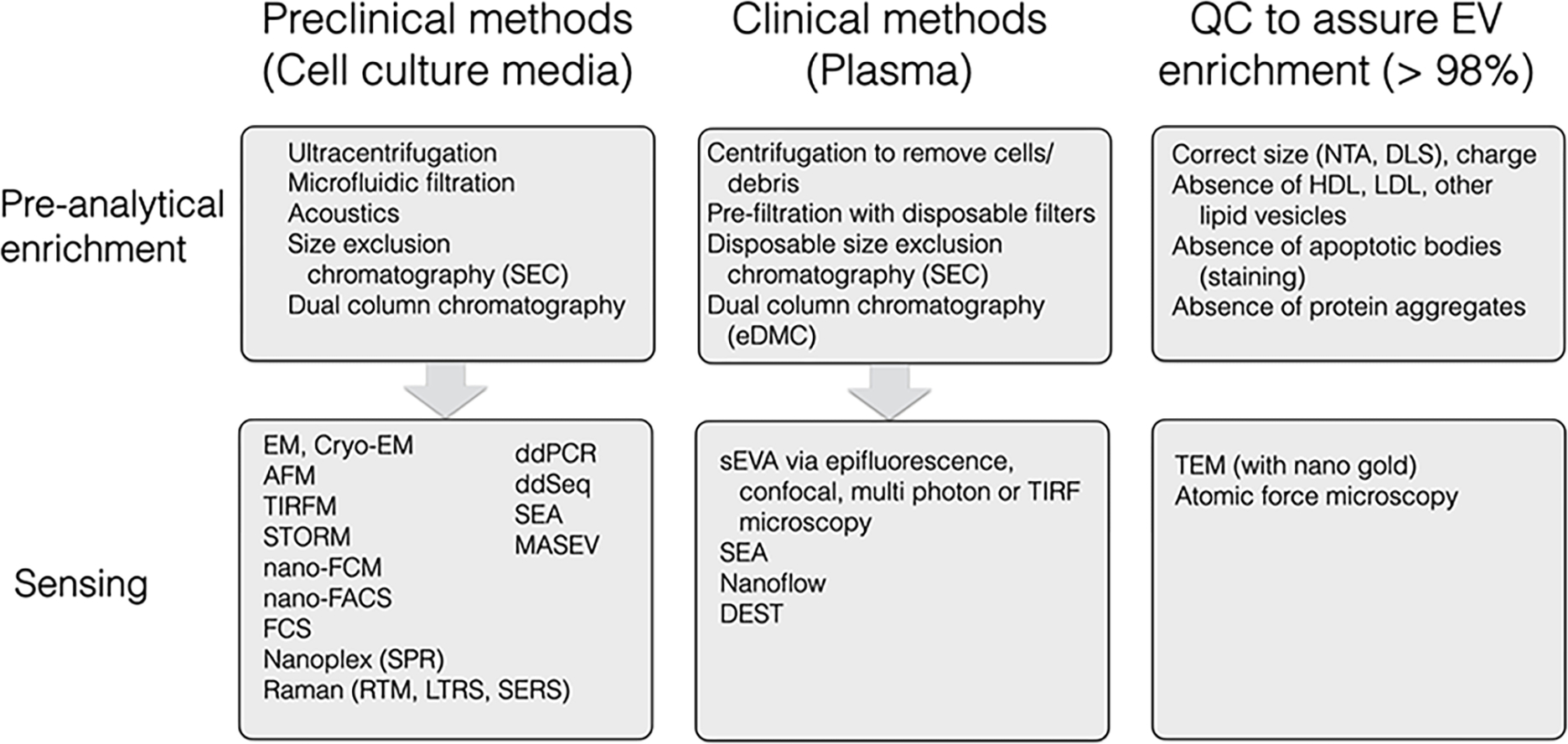 Figure 1. Summary of Individual EV Detection Methods
Figure 1. Summary of Individual EV Detection Methods
The authors mentioned that mutated KRAS protein, mutated P53 and other biomarkers were present in approximately 3% of EVs in patients with advanced pancreatic cancer and only 0.01% of EVs in patients with early-stage cancer. A clinical study indicated that current sEVA methods could detect approximately 90% of patients with stage 1 PDAC. With the ongoing development and advancement of clinically feasible single EV detection technologies, we are gaining a deeper understanding of the scarcity and complexity of ctEVs. Several questions are actively being explored: (i)How can existing single EV technologies be made more sensitive? (ii) Can multiplexing strategies be developed to interrogate ctEV to identify their organ of origin (to find small cancers in the human body); (iii)Is it possible to identify and differentiate ctEVs from relatively indolent (tumors doubling in years) versus highly aggressive (tumors doubling in months) cancers? We expect that the sensitivity of sEVA will increase with the emergence of improved antibodies against key targets in the future. Areas for improvement include more efficient pre-analysis techniques to prevent the loss of rare ctEVs, integration of sEVA with microfluidics, and enhancement of optical systems. Although methods like digital droplet PCR (ddPCR) and ddEV-seq have been developed, they are complex, time-consuming, and require further optimization. Another approach involves cycle imaging of different EV targets, such as using four channels (488 nm, 535 nm, 594 nm, 647 nm) imaged over five cycles to assess 20 biomarkers in a single EV.
Another area of significant clinical interest is combining of sEVA with cross-sectional imaging to achieve: (i) spatial localization of targets; (ii)temporal information provision; and (iii) improved differential diagnosis capabilities. Current diagnostic tools are lacking in effectively stratifying patients for the most suitable treatment, avoiding treatment-related toxicities, and achieving cost-effectiveness. Dynamic contrast-enhanced functional CT imaging (DCE-CT) is generally insufficient in this regard. sEVA holds promise in revolutionizing existing imaging methods by integrating EV biomarkers to enhance treatment monitoring and clinical decision-making, although substantial work remains in this area.
In summary, sEVA represents a promising frontier in research that will elucidate the biology of circulating EVs and, hopefully, facilitate the emergence of clinical trials for early detection across various malignancies.
Reference:
Ferguson S, Yang KS, Weissleder R. Single extracellular vesicle analysis for early cancer detection. Trends Mol Med. 2022 Aug;28(8):681-692. doi: 10.1016/j.molmed.2022.05.003. Epub 2022 May 24. PMID: 35624008; PMCID: PMC9339504.
Related Services:
