Tumor cell-derived extracellular vesicles (EVs) are being explored as circulating biomarkers for cancer detection. However, the effectiveness of bulk measurement in enabling early cancer detection remains uncertain. Professor Ralph Weissleder’s team from Harvard Medical School published an article in the journal Science Advances and found that single EV analysis (sEVA) technology can improve the accuracy of cancer diagnosis.

Pancreatic ductal adenocarcinoma (PDAC) is currently one of the leading causes of cancer-related mortality. Currently, surgical removal remains the sole curative option for pancreatic cancer, provided it is detected while still localized. Under the current treatment model, only 20% of patients are deemed operable, while 40% will progress to develop local but unresectable disease, and the remaining 40% will experience metastasis. Despite state-of-the-art computed tomography (CT), magnetic resonance imaging (MRI), and endoscopic ultrasound for patients with “imaging-detected localized disease” and surgery, the rates of positive resection margins range from 40% to 70%, and peripancreatic lymph node lesions exhibit a pathological positivity rate of 60% to 80%. To cure more patients with pancreatic cancer, new diagnostic tools will be needed to identify patients with preclinical and truly localized disease and to explore new treatments that allow for more R0/N0 resections.
High-resolution, contrast-enhanced CT and MRI imaging are commonly performed, but their sensitivity in detecting minimal disease is often limited, particularly in neoadjuvant and postoperative settings. Technological advances continue to rapidly improve the ability to detect circulating biomarkers in blood samples. These “liquid biopsy” markers include mutated or methylated cell-free DNA (cfDNA), tumor-associated extracellular vesicles (EVs), circulating tumor cells, and metabolic parameters (e.g., diabetes and muscle wasting). Although EVs, cfDNA, and metabolic markers have shown promise for PDAC diagnosis, further improvement in their current detection sensitivities is necessary for clinical utility. Particularly in early disease stages, lower diagnostic accuracy may stem from the substantial stromal component of PDAC, the coexistence of focal areas of pancreatitis indistinguishable from tumor-free pancreatitis, and the scarcity of early circulating biomarkers.
Researchers have developed many methods for analyzing EVs. Many early technologies relied on batch measurements, requiring 102 to 106 EVs for a single measurement. Although single EV methods (such as single EV analysis (sEVA) with multifluorescence, single particle interference reflection fluorescence imaging, nanoparticle tracking analysis, microfluidic resistive pulse sensing, and nanoflow cytometry) are actively developed, they measure different parameters, usually size rather than the ratio of molecular biomarkers. Regardless of the specific physical measurement, the next question concerns which molecular biomarker to analyze. Although research is ongoing, it is clear that many of the described biomarkers falter in larger clinical validation cohorts. Hence, a key question arises regarding whether this failure results from the selection of a given molecular biomarker, the extreme heterogeneity of PDAC, or the challenge of accurately detecting the very small number of biomarker-positive EVs within the normal EV population.
The authors believe that one way to address the current problem is to perform sEVA, as bulk methods may fail to identify small numbers of tumor-originated EVs, such as those found in microscopic cancers, within the context of host EVs. They further hypothesize that mutated oncoproteins (e.g., KRASmut) or tumor suppressor genes (e.g., P53mut) can be detected in individual EVs and that the same mutations occurring in cancer cells are responsible for increased tumor EV shedding. The choice to detect these mutant proteins is driven by their early appearance in the PDAC developmental cascade. Finally, the authors reason that detection efficiency could be enhanced by multiplexing measurements of several biomarkers. On the basis of these paradigms, a robust sEVA technology was developed and validated, allowing for multiplex protein measurements in single EVs. sEVA technology can answer the following questions: (i) What is the inherent heterogeneity of putative cancer cell-associated proteins in EVs? (ii) What is the frequency of mutated tumor/tumor suppressor proteins in individual EVs? (iii) What are the expression levels of two or more cancer-related proteins on a single EV. “Covering” more tumor-derived EVs (tEVs) through non-overlapping markers or obtaining “highly-specific” EV identifications (such as KRASmut and P53mut) are reliable options to improve clinical PDAC diagnosis.
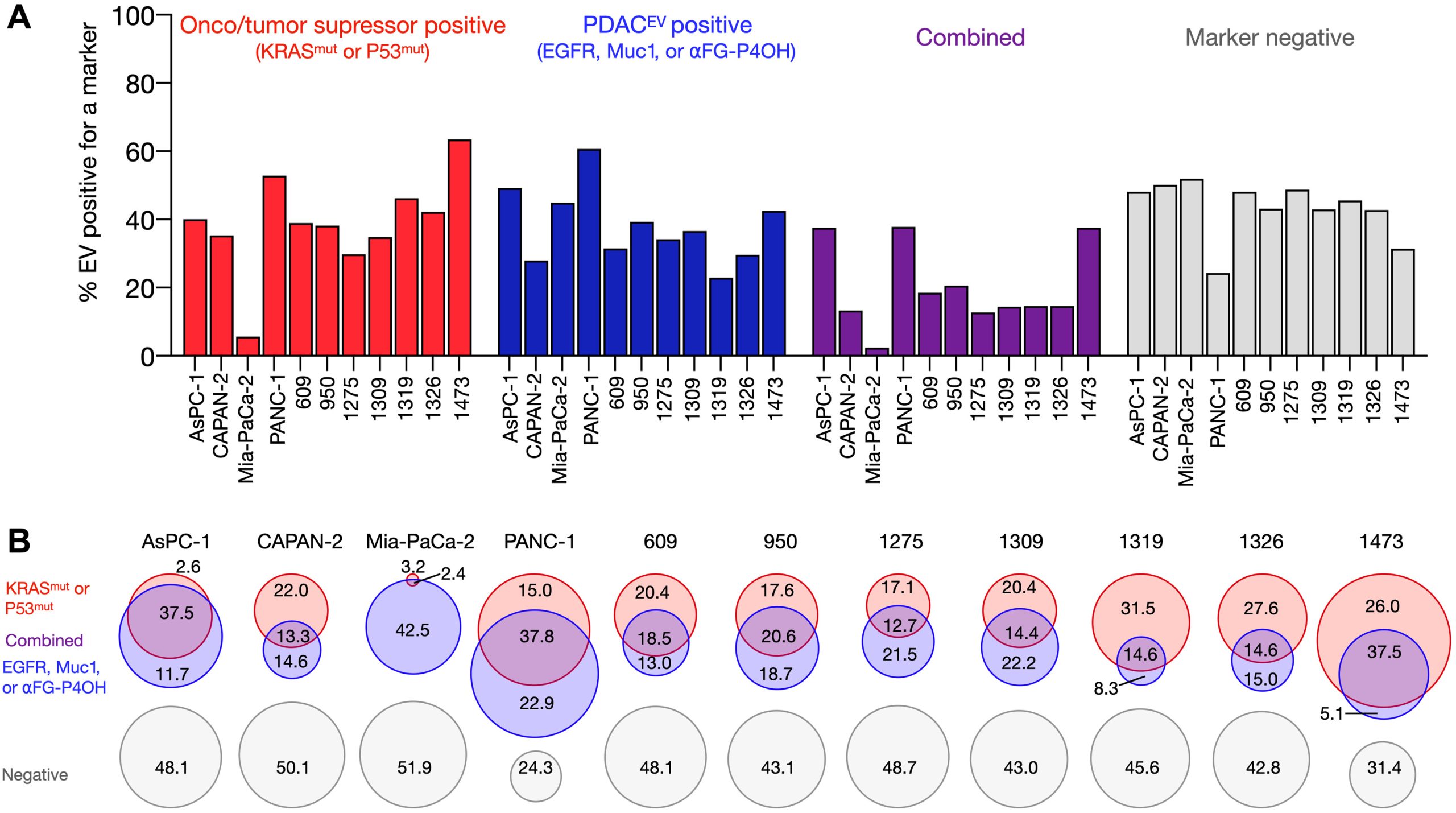 sEVA Analysis of EVs Derived from Multiple PDAC Cell Lines (Using pancreatic cancer (PDAC), the authors analyzed the composition of putative cancer markers in 11 cell lines. They observed that in parental PDAC cells expressing KRASmut and/or P53mut proteins, only approximately 40% of EVs exhibited the same positivity for these markers.)
sEVA Analysis of EVs Derived from Multiple PDAC Cell Lines (Using pancreatic cancer (PDAC), the authors analyzed the composition of putative cancer markers in 11 cell lines. They observed that in parental PDAC cells expressing KRASmut and/or P53mut proteins, only approximately 40% of EVs exhibited the same positivity for these markers.)
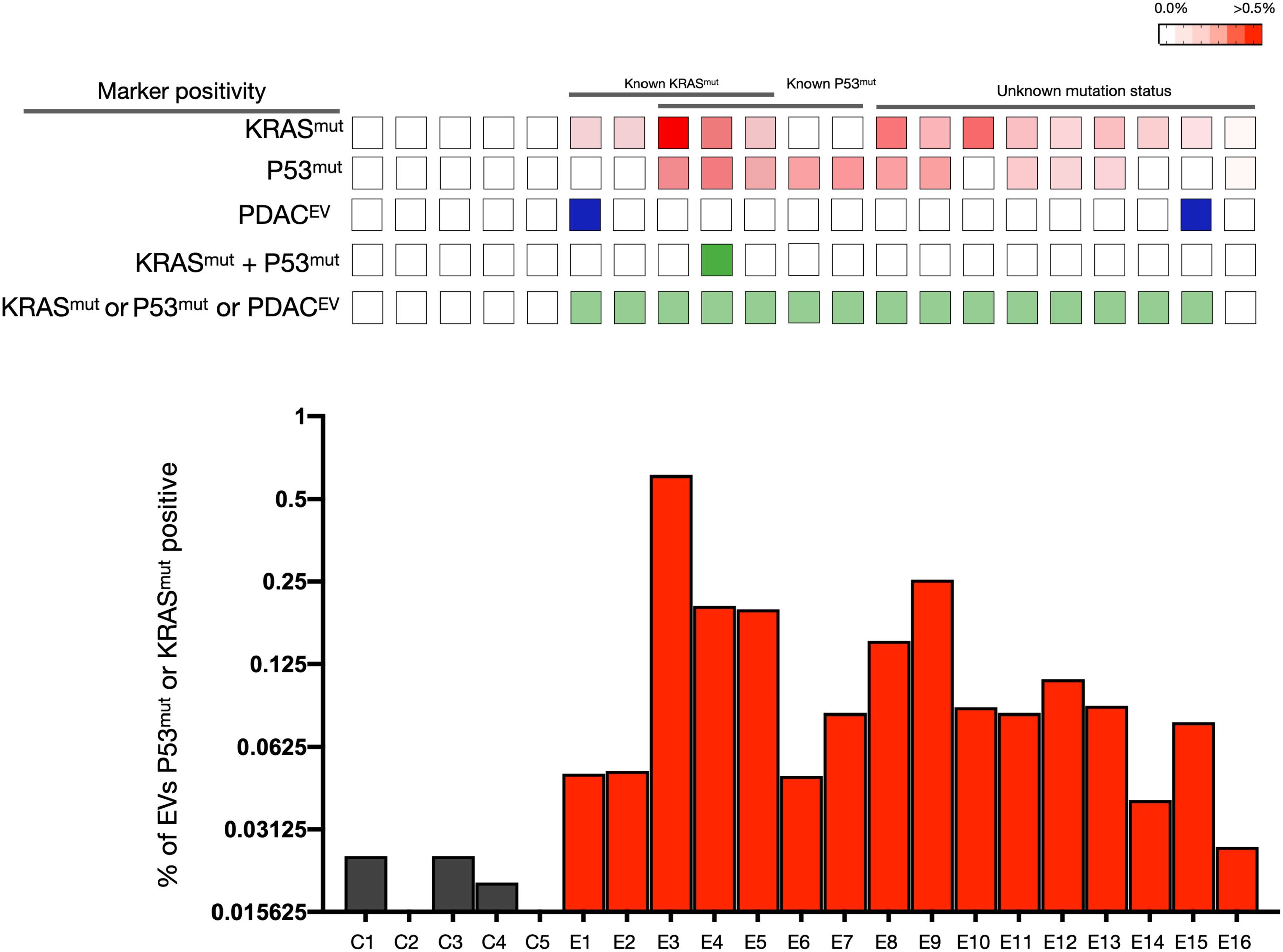 sEVA Analysis Results of Clinical Stage 1 PDAC Cases (A blinded study involving 16 patients with surgically confirmed stage 1 PDAC revealed that KRASmut and P53mut proteins were detectable at much lower levels, typically found in less than 0.1% of vesicles (i.e., positive in patient samples where the proportion of vesicles is usually less than 0.1%). These vesicles were detectable by the new sEVA method in 15 out of 16 patients.)
sEVA Analysis Results of Clinical Stage 1 PDAC Cases (A blinded study involving 16 patients with surgically confirmed stage 1 PDAC revealed that KRASmut and P53mut proteins were detectable at much lower levels, typically found in less than 0.1% of vesicles (i.e., positive in patient samples where the proportion of vesicles is usually less than 0.1%). These vesicles were detectable by the new sEVA method in 15 out of 16 patients.)
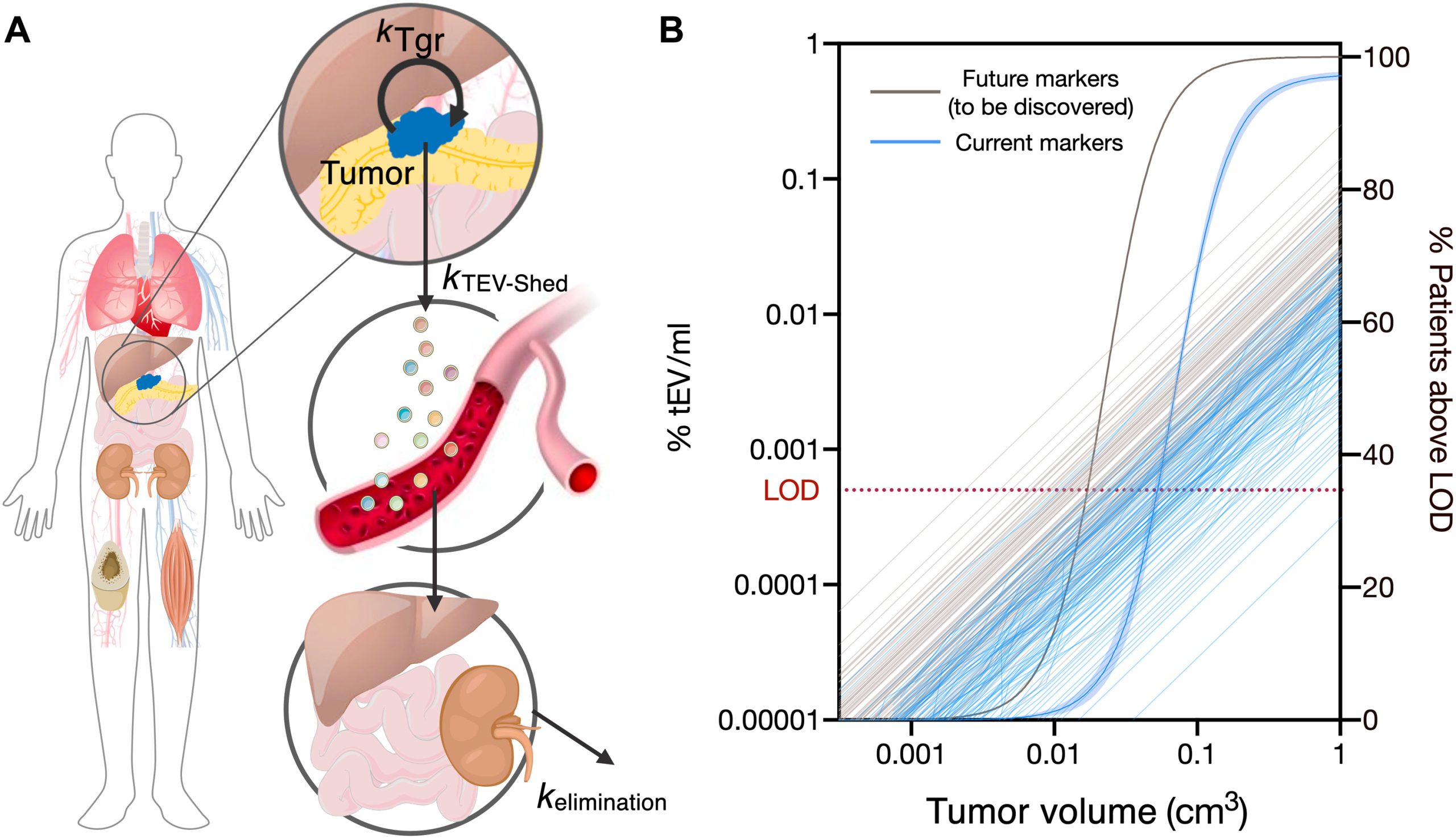 Modeling PDAC Cancer Detection Using Plasma sEVA (Utilizing a modeling approach, the authors estimated that the PDAC detection limit, based on single EV analysis (sEVA) technology, is approximately 0.1cm3 of tumor volume. This surpasses the capabilities of clinical imaging methods. These findings establish the potential of sEVA for early cancer detection.)
Modeling PDAC Cancer Detection Using Plasma sEVA (Utilizing a modeling approach, the authors estimated that the PDAC detection limit, based on single EV analysis (sEVA) technology, is approximately 0.1cm3 of tumor volume. This surpasses the capabilities of clinical imaging methods. These findings establish the potential of sEVA for early cancer detection.)
Reference:
Ferguson S, Yang KS, Zelga P, et al. Single-EV analysis (sEVA) of mutated proteins allows detection of stage 1 pancreatic cancer. Sci Adv. 2022;8(16):eabm3453. doi:10.1126/sciadv.abm3453
Related Services:
Pancreatic Cancer Tissue Exosome Research and Application
Therapeutic Exosomes for Pancreatic Cancer
Single-Cell & Tissue Exosome Combined Proteomics Research Service
