PreciAbTM
PreciAb™ Platform
Antibody Design
Antibody Structure Modelling
Antibody-Antigen Complex Analysis
Computer-aided Affnity Maturation
Antibody Structure Determination
Antibody Reformatting
Antibody to ScFv Reformat
Antibody to VHH Reformat
Antibody Developability Prediction
Antibody Aggregation Prediction
Antibody Immunongenicity Prediction
Antibody Screening & Design
Company
About Us
Contact Us
Downloads
Careers
Events

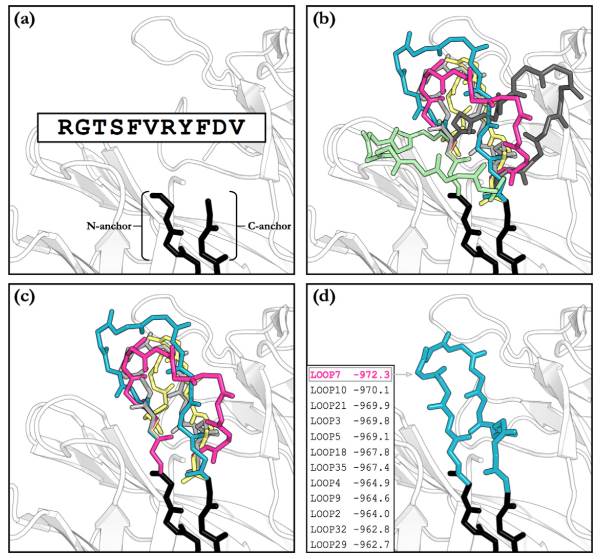 Fig.1 The major steps in an H3 loop modelling algorithm. Part A shows the inputs to the algorithm, which are an antibody structure with a missing loop, and the sequence of that loop. Part B shows the decoy generation, part C shows the filtering of structures, and part D shows the ranking and selection of the final prediction.1
Fig.1 The major steps in an H3 loop modelling algorithm. Part A shows the inputs to the algorithm, which are an antibody structure with a missing loop, and the sequence of that loop. Part B shows the decoy generation, part C shows the filtering of structures, and part D shows the ranking and selection of the final prediction.1

 More than 10 years of exploration and expansion
More than 10 years of exploration and expansion


