Before we start to introduce the principle of antibody diversity, you’d better learn about structure of antibody. Antibody consists of two heavy chains and two light chains. Most antibody related research is for human disease therapy, so here we only describe the principle of antibody diversity in human.
Source of Antibody Diversity
Antibody diversity in humans comes from several stages of Ig (Immunoglobulin) development, including both pre-immune repertoire, in which antibodies are developed for sampling the system for foreign bodies, and the post-immune repertoire, in which antibodies against foreign antigens are selected for and matured. In the pre-immune repertoire, there are four major sources of antibody diversity. The first source of pre-immune diversity is the recombination of VH, DH, and JH chains to build a functional VH chain, and VL and VJ (λ or κ) chains to build a functional light chain. To build a functional VH gene, there is a random assortment in which one of 39 functional VH genes is coupled with one of 27 functional D genes and 6 JH chains. Second, within the VDJ joining process, the junctional diversity created in the recombination process adds infinitely more diversity in 4 different ways. 1. The D genes can be translated in any one of three open reading frames in either direction to yield a total of six possible peptide fragments. 2. During the rearrangement process, a hairpin is formed that when resolved can result in the addition or deletion of N nucleotides resulting in added diversity. 3. About the VDJ joining mechanism, N nucleotides can be added or deleted as part of the VDJ joining process. In some cases, the coding sequences for several amino acid residues can be lost during the recombination process. 4. N nucleotides can be added or replaced by the activities of TdT (Terminal deoxynucleotidyl Transferase), particularly on either side of the D segment within the VDJ junctions, which make up CDR-H3 in the functional V-region. It has been estimated that these N changes can result in up to >107 different variations of CDR-H3, including CDR lengths ranging from just a few amino acid residues to over 25. Combined together, the somatic rearrangements, the combinatorial diversity within chains, the inherent mutagenesis that occurs in the assembly process, and the combination of heavy and light chains result in a potential pre-immune diversity of >1016 different antibodies.
After exposure of cell-bound IgM to antigen, the antibody genes undergo an affinity maturation process, generating new diversity from which antibodies with higher affinity to the targeted antigen epitope are selected, resulting in more effective binding and elimination of the antigen from circulation during the secondary immune response. It has been demonstrated that during the affinity maturation process, the average number of mutations in VH and VL are eight and five, respectively.
Human Antibody Gene Organization
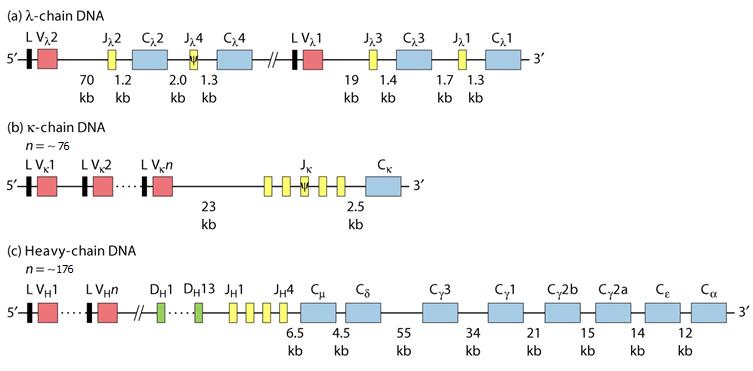
Antibody molecules are encoded by three independent groups of genes. Two genes dictate λ chains: one comprises Vλ and Cλ genes; k comprises Vk and Ck, genes, while the third group dictates H chains and has VH and CH genes.
The heavy chain variable region is itself composed of three segments, V (variable chain), D (diversity chain) and J (joining chain), while the light chain variable region has two segments, V and J. There are a total of 170–176 IGH (Immunoglobulin heavy chain) genes, of which 76–84 are thought to be functional. The functional genes, i.e. those that can be incorporated into functional Ig proteins, include 38–46 IGHV genes (39 is the number given by many sources) that fall into 6–7 families based on sequence similarity, 23 IGHD genes, 6 IGHJ genes, and 9 IGHC (Immunoglobulin heavy chain constant) genes.
The genetic locus which encodes the κ light-chain gene is located on the short arm of chromosome 2. The κ locus contains a total of 82 genes in two large clusters including 76 IGKV gene segments, 5 IGKJ segments, and a single unique IGKC gene, encoding the kappa constant domain. Only about 31 to 36 of the IGKV genes, which fall into six sequence-related groups, are considered to produce functional Vκ chains.
In distinct contrast to the κ genes, the λ-gene locus is comprised of multiple distinct Cλ regions, each of which may possess its own associated Jλ segment. These λ genes are tandemly arranged along the long arm of chromosome 22. The λ locus spans 1,050 kbp and has a total of 87–96 genes, including 73–74 IGVL genes, 7–11 IGLJ genes and 7–11 IGLC genes. Of these, it has been determined that 29–33 of the IGVL genes, which fall into 10 sequence groupings, can produce functional proteins. Additionally, 4–5 of the IGLJ genes and 4–5 of the IGLC genes are thought to be functional (Fig. 1).
Fig. 1 Organization of Ig (Immunoglobulin) gene
Antibody Gene Rearrangement and Diversity In Vivo
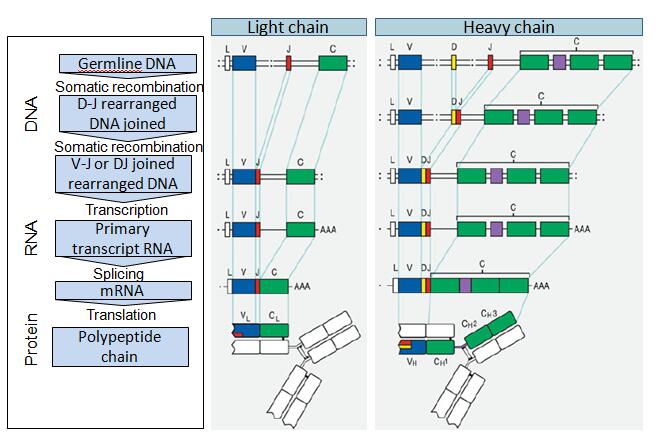
Before synthesizing Ig chains, however, Ig genes must be assembled within a genome of differentiating cells (rearrangement). The overall, high-level, assembly process for the heavy chain and light chain gene assembly and translation into an IgG antibody sequence is shown in Fig. 2. The extraordinary diversity of human antibodies comes from the combination of the assembly of different VH, DH, and JH, Vκ and Jκ segments, or Vλ and Jλ segments, and pairing of the heavy and light chains, to generate the primary antibody repertoire. The process of Ig diversification in humans is very complex and hierarchically coupled with the five stages of B cell development in the bone marrow, with each biochemical and genetic stage required for the next step and matched to a cell development stage.
Fig. 2 Antibody gene and protein assembly in vivo
Abbreviations: C: constant chain; D: diversity segment; J: joining chain; V: variable chain. Note that while only single V, J, D, and C regions are shown for simplicity, these are selected from a large number of genes. The major processes leading to the next step in antibody assembly are noted on the left. RNA transcripts are noted by “AAA” designating the poly-A tails.
The following antigen independent steps in B cell development occur in the bone marrow:
- Stem cell – heavy chain (IGH) and kappa (κ) and lambda (λ) light chain (IGK and IGL) genes are in germline configuration.
- Early pro-B cell – IGH undergoes D–J gene rearrangement with loss of DNA between the joined D and J segments. Upon induction of recombination activating gene 1 (RAG1), RAG2, and TdT in the pro-B cells, the DH and JH gene segments are joined first by a RAG-1, 2-dependent process.
- Late pro-B cell – IGH undergoes V–DJ rearrangement (VDJ rearrangement) with loss of DNA between the joined V and D segments. Recombination can be somewhat imprecise, since during this process several nucleotides may be removed from or added to a junction, so combination of VDJ segments of H chains results in a large number of possible sequences (that is, antibodies). This reshuffling process, known as combinatorial joining, is the prevalent source of Antibody
- Small pre-B cell – V–J rearrangement of light chain gene(s). κ chain is rearranged first then, if rearrangement of both κ alleles is unsuccessful, λ chain is rearranged.
- Large pre-B cell – intracellular expression and transient surface expression of m chain with invariant pseudo light chain. Successful cell surface expression of this receptor triggers allelic exclusion to prevent rearrangement of second allele and also initiates pre-B cell proliferation, which results in different light chains matched with the same heavy chain rearrangement in different daughter cells.
The following antigen dependent steps in B cell development take place in the periphery:
- Immature B cell – IgM surface expression. In the most common scenario, a VDJ segment joins first with Cμ genes, and subsequently with Cγ, Cε, Cα genes, with synthesis of a complete H IgM chain, etc. Without an associated L chain, surface expression is not possible and only cytoplasmic μ is found (pre-B cells).
- Mature naïve B cell – IgD and IgM expressed on cell surface, made from alternatively spliced transcripts.
- Lymphoblast – alternative splicing results in secreted IgM.
- Memory B cell – isotype switch to IgG. Somatic hypermutation of IGH occurs in the germinal center of lymph nodes. Mutated Ig are selected for improved antigen binding in a process termed affinity maturation.
- Subset of B cell expressing surface IgD and IgM undergo a further DNA rearrangement, class switch, which results in expression of IgG, IgA, or IgE, changing the IgH constant region, and therefore effector functions associated with the Ig, without changing specificity
- A subset of B cells survive as long-lived memory B cells, which respond rapidly to antigen and differentiate into antibody producing plasma cells.
- Terminally differentiated plasma cell – alternative splicing yields both membrane-bound and secreted Ig.
Gene rearrangement mechanism is detailed here:
VDJ recombination proceeds via precise DNA cleavage initiated by the RAG proteins (RAG-1 and RAG-2) at short conserved signal sequences [128]. Whatever their precise role, the coordinated expression in pre-B is essential for the rearrangement of Ig genes, but RAG activity is switched off in mature lymphocytes. Rearrangements are carefully orchestrated, following the principle of allelic exclusion, that is, in each B cell is transcribed the gene product of only one of each chromosome pair. The gene located on the second chromosome typically is not used, to prevent contemporary synthesis of H chains with differing V domains in any given cell; it is an allele, a term designating two genes or two or more alternate forms of a single gene occupying the same locus. The gene on the second chromosome is not rearranged unless a nonproductive rearrangement occurs; when rearrangements are nonproductive on both chromosomes, cell death ensues, thereby by clarifying why the same B lymphocytes within their entire life span can produce only a single type of L chain (κ or λ). Ig V, D, and J gene segments are flanked by conserved recombination signal sequences (RSS), consisting of a heptamer and a nonamer separated by a non conserved spacer of either 12 or 23 nucleotides. The 12–23 base pair rule first postulated to explain Ig gene rearrangement. Virtually, during a rearrangement, a gene with a flanking sequence containing a 12-base pair spacer can only join to a gene whose flanking sequence has a 23-base pair spacer and vice versa, thereby elucidating the precise order of Ig gene transcriptions. Firstly, RAG-1 recognizes RSS and form complex of RAG-1/RAG-2 which cut single sequence of DNA. And then a hairpin is formed by addition of copies of the last nucleotides of the coding region (templated or “P” additions) or by random nucleotide additions by the enzyme TdT (TdT, “N” additions). The joining of the coding ends of the rearranged gene segments is imprecise due to base additions, base losses and out of frame joining. This imprecision of joining generates “junctional” diversity. There is similar gene rearrangement mechanism in T cells (T lymphocytes).
Principle of antibody diversity is completely understood. There is very close relationship in amino acide sequence and antibody function. As we know, there is huge diverse function between two antibodies which have almost same amino acid even if one amino acide is different. Based on it, accuracy of antibody sequencing is very important in related research. Creative Biolabs Provides world-class de novo antibody sequencing services with 100% accuracy for research, diagnostic, and therapeutic industries.
Reference: Anne Davidson. The Role of Variable Region Gene Rearrangements in the Generation of Autoantibodies. Contemporary Immunology. pp 177-19.