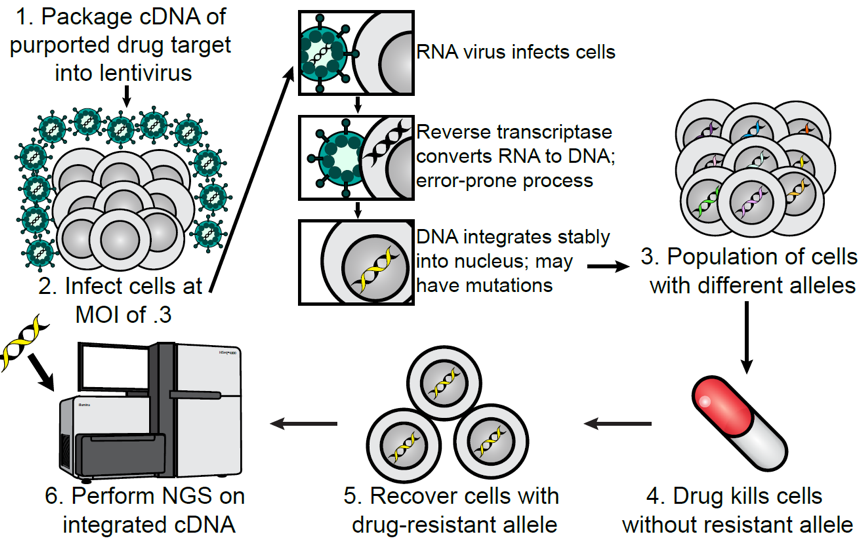
The identification of drug resistance mutations in drug targets provides vital information for new drug development. Due to the fallibility of HIV-1 reverse transcriptase (RT), lentivirus transduction can lead to many types of mutations. The researchers optimized and used this feature to identify drug resistance mutations and developed a technique called LentiMutate. This technique was validated by identifying clinically related EGFR resistance mutations and then applied to two other clinical anticancer drugs: bcr-abl inhibitor imatinib and KRAS G12C inhibitor AMG510. The novel deletions of bcr-abl1 results in resistance to imatinib. In KRAS-G12C or wild-type KRAS, point mutations in the AMG 510 binding pocket or oncogenic non-G12C mutations can lead to resistance to AMG510. LentiMutate has been proved to be of high value in clinical and preclinical cancer drug development.
By referring to drug-resistant mutations that may be clinically relevant, and information about the target and the chemical structure of the drug, the mutation of the drug target confirms the its activity and helps develop new drugs that circumvent clinical drug resistance. Therefore, identifying such mutations that lead to drug resistance can provide crucial information for clinical and preclinical drug development projects. Multiple techniques, such as in vitro error-prone PCR, plasmid mutagenesis in error-prone DNA replication bacteria, ENU mutagenesis, the use of mismatch repair defective cell lines, and mutagenesis by integrated TilEs followed by sequencing (MITE-SEQ), have been deployed to identify mutations associated with drug resistance. All of these techniques are limited by identifying only point mutations, challenges in sequencing, or constraints in potential sequence space, despite their respective pros and cons. In addition, these technologies can be technically challenging, laborious, and costly.
Overview of LentiMutate. (Cancer Research)
Since lentivirus transduction has been routinely carried out in many laboratories, it is relatively simple to use LentiMutate. The researchers envisioned that LentiMutate could be used in the early stages of cancer drug discovery to test specificity, provide information for the synthesis of second-generation inhibitors, and improve the understanding of drug-protein interactions. It is worth noting that LentiMutate can be used in any type of RNA/DNA/protein evolution process, with positive selection steps, not just the identification of drug resistance mutations. Therefore, the use of LentiMutate is not limited to providing information for anti-cancer drug development projects.