In a groundbreaking study, researchers from the Genome Institute of Singapore (GIS) developed a new computer model of machine learning (an artificial intelligence model, AI) to accurately find tumor mutations. They also discovered that a new gene mutation in noncoding DNA may have caused gastric cancer. The new technology developed in this study will help researchers explore the effects of non-coding DNA mutations in other tumors in the future.
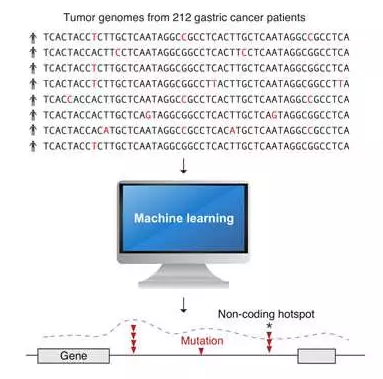
Photo Credit: Agency for Science, Technology and Research (A*STAR), Singapore
Cancer is one of the biggest causes of death in the world, resulting from abnormal cell growth due to DNA mutations, and stomach cancer is the fourth most deadly cancer in the world. The 2% of the coding DNA in our genome has been fully studied, but the other 98% of non-coding DNA is still in an unknown state. Non-coding DNA can regulate gene activity, and more and more pieces of evidence show that gene mutations in these gene regions also contribute to the development of cancer.
In this study, the researchers created two AI methods to scan the entire genome of 212 gastric cancer tissues, which takes 30 years to finish the analysis by an existing standard model computer. Using computer groups in GIS and National Supercomputing Centre Singapore (NSCC), the researchers successfully identified several new cancer-related gene mutations in the genome. The analysis also showed that non-coding DNA mutations can be caused by changing the three-dimensional structure of the genome.
Dr. Anders Skanderup, a GIS researcher and lead scientist for the study, said:”We focused on using computational and data-driven methods to study the root causes of cancer to help develop more effective strategies. Our analysis showed that 11 mutations non-coding genes could regulate the three-dimensional structure of the genome, and almost one in four gastric cancer patients had mutations in these areas.”
He continued: “These non-coding gene mutations are also found in other cancers such as colorectal cancer, pancreatic cancer, and liver cancer. Therefore, these gene mutations can serve as biomarkers for detecting and monitoring the progression of these diseases.”
Professor Ng Huck Hui, managing director of GIS, said:”Past studies have only focused on searching for coded DNA that only accounts for 2% of our genome. Therefore, there has been a question for many years whether we missed important information by ignoring the remaining 98%. This is the first study about the effect of non-coding DNA on gastric cancer. We hope it can motivate more researchers to participate in similar studies to reveal the mechanisms and effects of these specific mutations.”
Reference:
Yu Amanda Guo et al. Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers, Nature Communications (2018). DOI: 10.1038/s41467-018-03828-2
