iPSC Reprogramming by mRNA: A Safe and Efficient Method
Reprogramming iPSC using synthetic mRNA involves the delivery of key transcription factors (e.g., ECT4, SOX2, KLF4, c-MYC), which are transformed into somatic cells to restore them to pluripotency. This approach avoids genomic integration and eliminates risks. Insert mutations and ensure that the "footprint-free" iPSC has a high degree of genomic integrity.
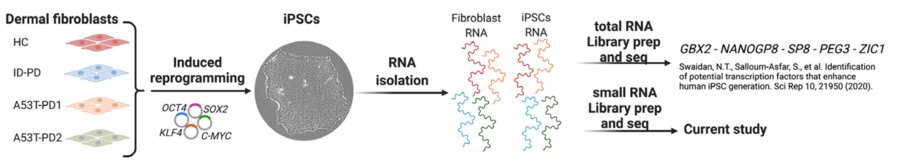 Fig. 1 Sendai virus-mediated reprogramming of human fibroblasts into iPSCs.1,4.
Fig. 1 Sendai virus-mediated reprogramming of human fibroblasts into iPSCs.1,4.
Service Advantages
- Non-integrating & genomically safe - RNA reprogramming avoids viral or DNA-based integration, ensures no risk of mutagenic and complies.
- High efficiency & speed - The optimized protocol achieves more than 80% reprogramming efficiency in 12-16 days, significantly faster than traditional virus methods.
- Customizable protocols - Tailor mRNA cocktails (e.g., OSKM, M3O fusion factors) and delivery methods (lipid nanoparticles, electroporation) to suit cell type and project needs.
- Robust quality control - Stringent validation of pluripotency markers (TRA-1-60, SSEA4), karyotyping, and functional assays ensures cell authenticity and performance.
- Scalability - Generate iPSCs from small research batches to large quantities, with seamless integration into downstream workflows (e.g., differentiation, cryopreservation).
Technical Process
| Process | Description |
|---|---|
| Cell Preparation | Isolate and culture somatic cells (fibroblasts, PBMCs, etc.) using feeder-free, xeno-free media. |
| mRNA Design & Synthesis | Custom mRNA sequences encoding reprogramming factors (Oct4, Socx2, Klf4, c-Myc) has modified nucleic acid to enhance stability and reduce immunogenicity. |
| Transfection | Delivery of mRNA via lipid nanoparticles or electroporation optimized for cell-type specific uptake. |
| Reprogramming Activation | Sequential injection to maintain pluripotency factor expression. |
| Colony Formation & Validation | Monitor alkaline phosphatase activity and pluripotency markers (Oct4, Nanog). |
| Expansion & Cryopreservation | Scale iPSC colonies and bank cells under liquid nitrogen for long-term storage. |
Cutting-Edge Technologies Driving Our Services
To ensure the highest standards of safety, efficiency, and translational relevance, our iPSC reprogramming service integrates the following advanced methodologies.
- Self-Replicating RNA (srRNA): Single infection reprogramming using synthetic SrRNA constructs to maintain pluripotency factor expression, reduce cellular stress and increase efficiency.
- Optimized Lipid Nanoparticles (LNPs): Enhance mRNA stability and cell uptake with proprietary LNP formulation, achieving high efficiency in different cell types.
- High-Resolution Genomic Surveillance: Genome-wide SNP/CNV analysis ensures that iPSC lines have no insertion mutations or chromosomal abnormalities.
- MicroRNA-Augmented Cocktail: Fusion mRNA-367/302 simulates accelerated reprogramming kinetics and improved differentiation potential.
- Automated Epigenetic Workflows: Artificial intelligence-driven epigenetic reshaping ensures expression of homogeneous pluripotency markers (OSC4, NANOG) and minimizes batch variability.
- Multi-Omics Quality Control: Single-cell RNA sequencing and proteomic analysis verify the homogeneity and functional authenticity of iPSC.
Applications of iPSC Reprogramming
Table 1 Why choose mRNA reprogramming?
| Feature | mRNA-Based | Traditional Methods |
|---|---|---|
| Safety | No genomic integration | Risk of insertional mutagenesis |
| Efficiency | Up to 20% efficiency | 0.1–1% efficiency |
| Timeline | 12–16 days | 3–4 weeks |
| Customization | Fully adaptable protocols | Limited to fixed viral vectors |
| Clinical Suitability | GMP-compliant, mutation-free | Requires extensive safety validation |
Our applications of iPSC include but not limited to:
- Disease Modeling - Generate iPSCs from patients with genetic disorders (e.g., Parkinson's, ALS) for pathogenesis studies.
- Drug Screening - Test compound toxicity or efficacy using iPSC-derived cardiomyocytes, hepatocytes, or neurons.
- Regenerative Medicine - Develop cell therapies for neurodegenerative diseases, diabetes, or cardiovascular repair.
- Basic Research - Investigate developmental pathways, epigenetic reprogramming, and cell fate determination.
Published Data
The researchers focused on screening protein-coding genes involved in iPSC reprogramming. This screen identifies 24 reprogramming roadblock genes. Of those, KO of the previously uncharacterized mouse KRAB-ZFP gene Zfp266 accelerates the kinetics of reprogramming and improves the efficiency of iPSC generation by 4- to 10- fold in various reprogramming contexts. This work serves as a resource for better understanding reprogramming mechanisms and highlights SINEs as a previously undescribed TE class involved in pluripotency induction.
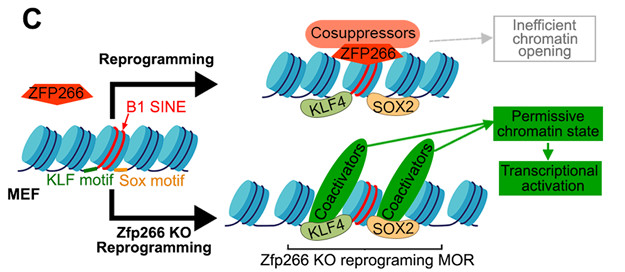 Fig. 2 Mechanistic model depicting how Zfp266 KO enhances reprogramming.2,4
Fig. 2 Mechanistic model depicting how Zfp266 KO enhances reprogramming.2,4
In this study, researchers examined the molecular mechanism for MET, focusing on RNA metabolism. DDX6, an RNA helicase, was indispensable for iPSC formation, in addition to RO60 and RNY1, a non-coding RNA, which form complexes involved in intracellular nucleotide sensing. The abrogation of DDX6 expression inhibited iPSC generation, which was mediated by RNA decay targeting parental mRNAs supporting mesenchymal phenotypes, along with microRNAs, such as miR-302b-3p. These results show that parental mRNA clearance is a prerequisite for cellular reprogramming and that DDX6 plays a central role in this process.
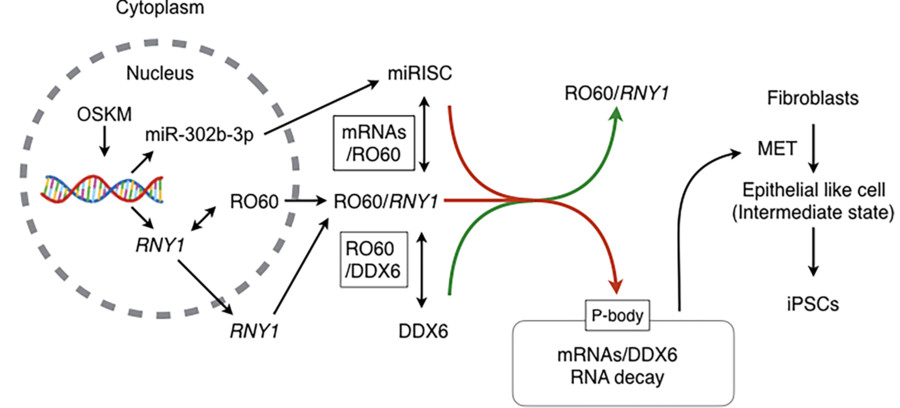 Fig. 3 Illustration of the proposed model to explain the molecular mechanism for RNA decay at the early reprogramming stage.3,4
Fig. 3 Illustration of the proposed model to explain the molecular mechanism for RNA decay at the early reprogramming stage.3,4
Client Success Stories
"Their mRNA reprogrammed iPSC allows us to model a rare neurodegenerative disease with unprecedented accuracy. These cells show true neuronal morphology and electrophysiological activity."
— Dr. Sarah Lin, Neurodegenerative Disease Researcher
"Compared to viral methods, the rapid reprogramming process saved us 6 weeks and accelerated our drug discovery pipeline."
— Dr. Alex Carter, Drug Development Lead
"Using mRNA reprogrammed iPSC-derived dopaminergic neurons, we have discovered a novel inhibitor that reduces a-synuclein aggregation. In our model of Parkinson's disease, there was a 75% drop."
— Dr. Elena Rossi, Senior Scientist, Parkinson's Research Institute
"For our diabetes research, their mRNA-reprogrammed iPSCs differentiated into beta cells with 92% purity (vs. 65% with Sendai virus)."
— Prof. Liam O'Connor, Stem Cell Biologist, University of Cambridge
FAQs
Q: Can I use mRNA reprogramming for primary cells with low proliferation rates?
A: Yes. Our protocols include optimized transfection enhancers and feeder-free media to support challenging cell types.
Q: Are your iPSCs suitable for 3D organoid cultures?
A: Absolutely. Our iPSCs maintain high pluripotency and differentiation potential for complex 3D systems.
Q: Do you offer CRISPR-edited iPSC lines?
A: Yes. We provide isogenic edited lines with precise mutations for disease modeling.
Q: Can I request custom epigenetic modifications during reprogramming?
A: Yes. Our platform introduces epigenetic markers (for example, H3K27ac activation, DNA demethylation) to achieve targeted chromatin remodeling at specific sites to enhance differentiation potential.
Take the Next Step with Creative Biolabs














1. Contact Us
via the Inquiry Form or Email
2. Define Your Needs
Cell Type, Function, Quantity, Modifications
3. Kickstart the Project
Our Expert Team Guiding Every Step
Our mRNA reprogramming service combines scientific rigor with industry-leading efficiency. Whether you need patient-specific iPSCs for disease research or scalable cell banks for drug development, we deliver solutions tailored to your goals.
References
- Salloum-Asfar, Salam, et al. "Combined noncoding RNA-mRNA regulomics signature in reprogramming and pluripotency in iPSCs." Cells 11.23 (2022): 3833. https://doi.org/10.3390/cells11233833
- Kaemena, Daniel F., et al. "B1 SINE-binding ZFP266 impedes mouse iPSC generation through suppression of chromatin opening mediated by reprogramming factors." Nature communications 14.1 (2023): 488. https://doi.org/10.1038/s41467-023-36097-9
- Kami, Daisuke, et al. "The DEAD-box RNA-binding protein DDX6 regulates parental RNA decay for cellular reprogramming to pluripotency." PLoS One 13.10 (2018): e0203708. https://doi.org/10.1371/journal.pone.0203708
- Distributed under Open Access license CC BY 4.0, without modification.
Created September 2025