Predictive Computational Modeling of Gut Microbiome
The gut microbiota has profound effects on our health, a realization leading to a surge of animal experiments in this area, so the development of a computer simulation tool to reduce or replace these animal experiments is a gap that is becoming increasingly urgent to fill. Creative Biolabs has developed a predictive computational platform by integrating novel state-of-the-art computational techniques and modeling approaches. Through computational models, new information can be leveraged to predict the relationship between gut microbiota and diseases.
Background
Microbiome data is especially enticing in human health research as it has the potential to explain medical mysteries that current clinical information has not been able to resolve. The advancements in the microbiome data sequencing techniques initiate the research on the microbiome and its relationship with the host organism. In detail, next-generation sequencing (NGS) has revolutionized the human gut microbiome. This also supports the most traditional metagenomic technique based on 16sRNA gene amplification via polymerase chain reaction (PCR) and whole-genome sequencing (WGS).
The development of sophisticated bioinformatics and data science tools for the analysis of large amounts of data followed. Coupling microbiome technologies, statistical and computational analyses, and more advanced disease models, these approaches promisingly provide the opportunity to analyze of community dynamic of gut microbiota and establish causal links between the microbiome and disease, and subsequently inform therapeutic development.
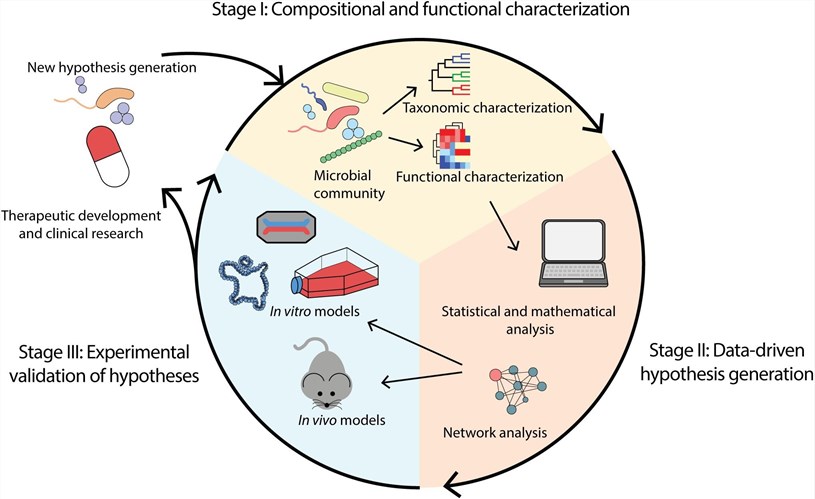 Fig.1 Overview of the microbiome analysis workflow.1
Fig.1 Overview of the microbiome analysis workflow.1
Services
As the microbiome field matures, more complex study designs have been used to study the human microbiome in various clinical and biological variables. Building upon the increased experimental data generated through the high-throughput approaches, computational modeling approaches have been developed to integrate and analyze data to study function. The computational tools can predict how the gut microbiome changes over time. We have the ability to a high level of computational and bioinformatics knowledge to establish models thus retrieving meaningful information from a large amount of multi-omics data. These methods involve machine learning methods to predict host health status based on the community taxonomical and community functional profiles, and then predict the ecological niche of bacteria.
A well-designed and validated gut-microbiota model could be used to study the dynamics of many hypothetical situations before clinical study. Here are some mathematical modeling approaches applied to the human gut microbiota.
Genome-scale metabolic models (GEMs)
GEMs are utilized to study various aspects of the gut microbiome including microbe-microbe, host-microbe and diet-microbe metabolic interactions.
Metabolic models
The flow of metabolites through each reaction in each species is determined such that an objective function (for example, biomass production) is optimized
Generalized Lotka-Volterra model
The change of species abundances over time is described at the level of populations.
Kinetic models
The change of species abundances and concentrations of key metabolites over time is described.
Network (topological model)
Species are represented as nodes, and their interactions as directed or undirected edges.
Neutral model
The change of species abundances over time is described at the level of individuals.
Computational modeling can help us to:
-
Improve our understanding of the gut microbiome
-
Work towards structure-based models that predict biotransformation of chemicals
-
Contribute to the functional annotation of microbiome sequencing data
-
Better understand metabolic host-microbiota interactions
Much of the effort in modern microbiome research focuses on generating multiple types of omic data. Creative Biolabs develops computational modeling analysis for integrating these multiple meta-omic datasets. Our microbiome services experts are available to guide predictive computational modeling of the gut microbiome. If you are interested in our service, please contact us for further information.
Reference
-
Young, Remy B., et al. "Key technologies for progressing discovery of microbiome-based medicines." Frontiers in microbiology 12 (2021): 685935. Distributed under Open Access license CC BY 4.0, without modification.
For Research Use Only | Not For Clinical Use


 Fig.1 Overview of the microbiome analysis workflow.1
Fig.1 Overview of the microbiome analysis workflow.1
 Download our brochure
Download our brochure