Peptide Identification Service
Peptide identification can be used to determine the accurate identity of proteins and to ascertain the expression patterns in specific organisms, tissues, or cells, as well as detailed information about antibodies, drugs, or other proteins. At Creative Biolabs, we utilize state-of-the-art tandem mass spectrometry (MS/MS) technology to identify and visualize the entire peptide repertoire of the clients' interests.
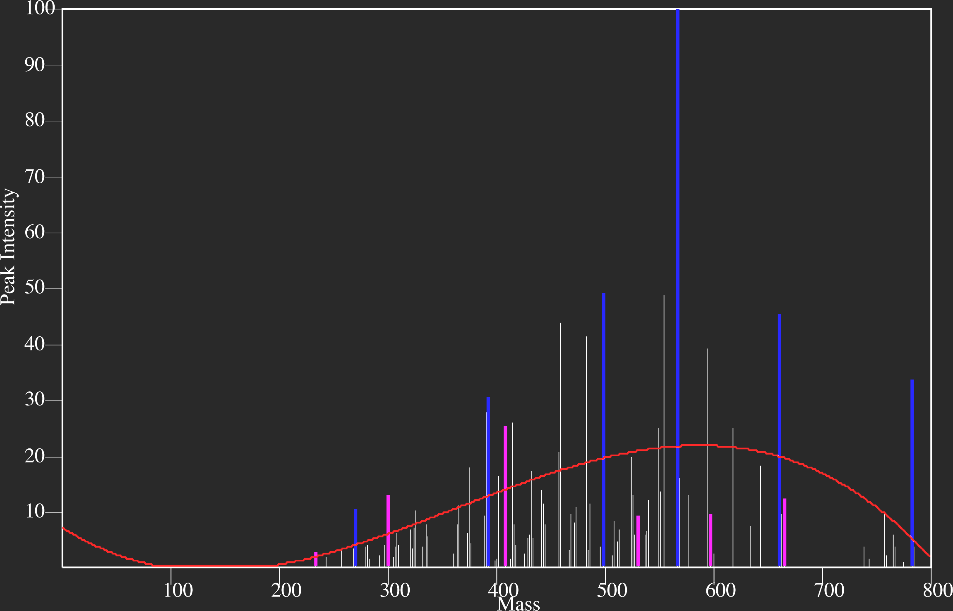 Fig 1. MS/MS data set for peptide identification.1
Fig 1. MS/MS data set for peptide identification.1
Peptide Identification Protocol
Once samples are received, peptides will be extracted from any biological sample submitted according to the specifications and separated into less complex peptide samples by LC. The separated samples will be ionized and detected using an MC. The ions of interest will be fragmented into product ions as precursor ions and undergo secondary mass spectrometry detection, generating an MS/MS spectrum.
Sample Submission
Generally, we require a sufficient sample quantity to extract enough valid information, but we are also willing to analyze low-concentration samples. We recommend submitting the following types of samples, and for other sample types, please contact our technical team.
Gel Samples
Please ensure thorough cleaning of gel samples and prepare clean containers to avoid potential contamination. You can submit cut strips or entire strips with annotated images to help us understand your requirements. Additionally, information about the dyes and reagents used during sample preparation is also required.
Pull-Down Assay
We require a minimum of 10 μg of protein to perform the digestion process, which is not limited to protein samples attached to beads or eluted. For all cases, we need you to provide information on antibodies, relevant reagents, and additional samples for negative controls.
Solution Samples
We may require a detailed information about buffer composition and concentration when submitting solution samples, and to avoid the presence of glycerol, PEG.
Results Output
Your samples will be carefully handled and analyzed, and the obtained MS/MS spectra will be systematically analyzed and computed to derive peptide identification information for your samples. The fragment spectra of the samples will be compared against existing databases to infer correct and plausible peptide sequences. In most cases, you will receive:
- Detailed explanations of experimental steps and mass spectrometry parameters
- Mass spectrometry analysis images of the sample
- Detailed information on peptide or protein identification
If we are unable to find the sequence you are testing for in the existing database, de novo sequencing is also an optional solution.
Our Services
As a renowned biotechnology company, Creative Biolabs specializes in protein/peptide identification, and employs various MS/MS fragmentation techniques for peptide sequencing. Whether you are aiming to identify naturally occurring peptides for proteomic research or utilizing our services for protein identification in proteomics, our experienced team of scientists and technicians is committed to delivering high-quality and reliable results. Please feel free to contact us to discuss any specific requirements.
Reference
- Gallia, J.; et al. Filtering of MS/MS data for peptide identification. BMC Genomics. 2013, 14: S2.