Intratumoral Microbiota Single-Cell Analysis Service
Background Service Workflow Advantages FAQs Contact Us
The tumor microenvironment is a complex ecosystem, and understanding its intricate components is crucial for advancing cancer therapies. To address challenges such as long drug development cycles, difficulty in obtaining high-quality recombinant proteins, and complexities in clinical trials, Creative Biolabs provides a cost-effective intratumoral microbiota single-cell analysis service. This service helps you accelerate drug discovery, develop highly specific antibodies, and streamline clinical trial processes through advanced single-cell sequencing and innovative bioinformatics techniques.
The Crucial Role of Intratumoral Microbiota in Cancer and Gene Therapy
Recent scientific insights have illuminated the critical role of intratumoral microbiota in influencing cancer progression, therapeutic response, and overall patient outcomes. These microbial communities, once thought to be absent, are now recognized as integral components of the tumor microenvironment (TME), impacting everything from immune modulation to genomic stability. While gene therapy holds immense promise for targeted cancer treatment, its efficacy can be significantly influenced by the local microbial landscape. Understanding the specific microbial populations at a single-cell resolution within tumors is paramount to unlock new diagnostic biomarkers, predict treatment responses, and design more effective, microbiota-informed gene therapies. This precision is essential for overcoming current therapeutic limitations and moving towards personalized oncology.
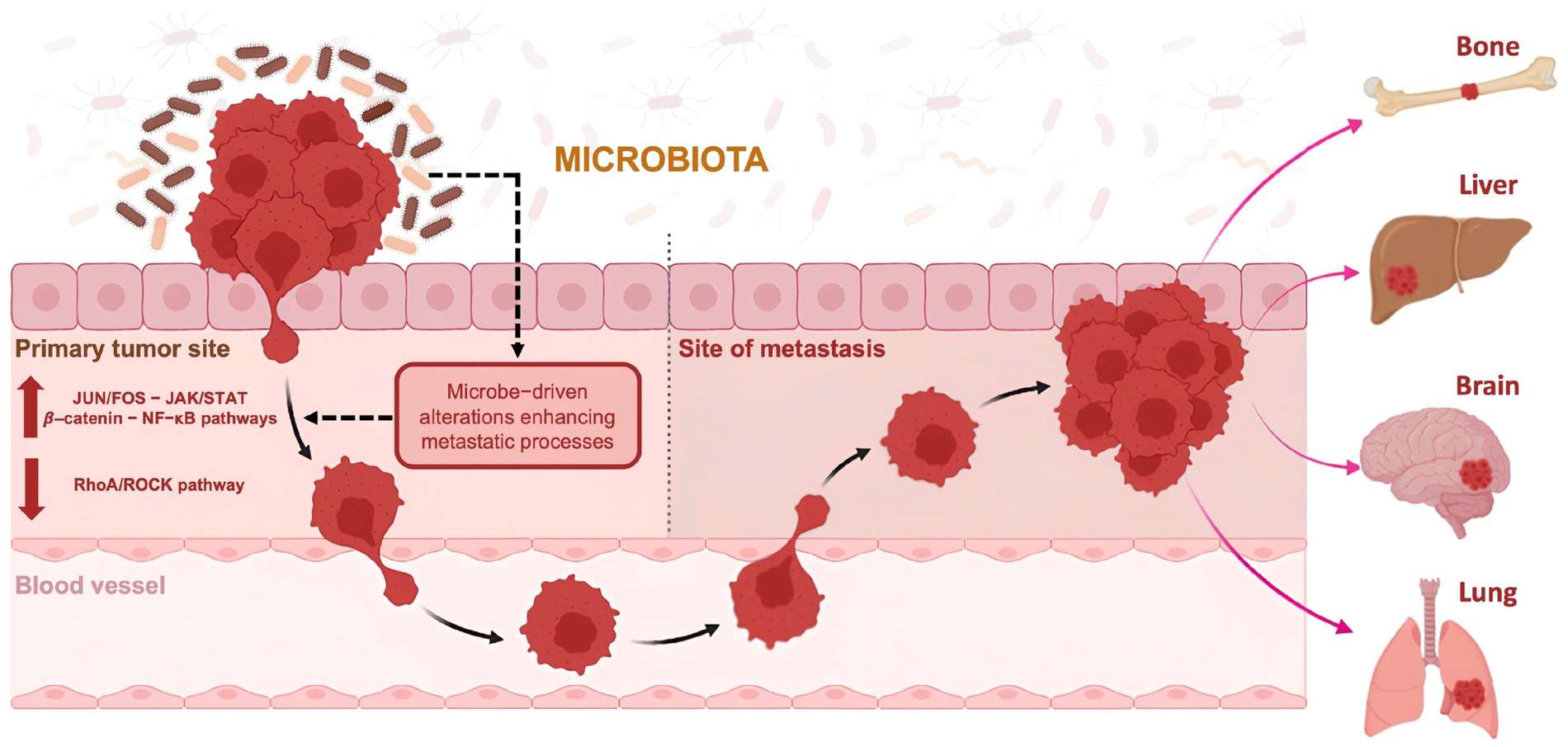 Fig.1 Intratumoral microbes can promote metastasis formation in cancer cells.1
Fig.1 Intratumoral microbes can promote metastasis formation in cancer cells.1
Intratumoral Microbiota Single-Cell Analysis Service at Creative Biolabs
Creative Biolabs' intratumoral microbiota single-cell analysis service provides unparalleled insights into tumor tissue microbial landscapes. Our assay utilizes advanced single-cell sequencing: isolating microbial and host cells from tumor samples, then extracting, amplifying, and sequencing their nucleic acids at high resolution for precise detection and characterization of microbial DNA/RNA within specific host cell populations. Analysis typically takes 8-14 weeks, from sample receipt to final report, varying with project complexity and volume, ensuring timely insights.
Workflow
Step 1: Sample Dissociation and Single-Cell Isolation
Tumor tissues are carefully dissociated to obtain a single-cell suspension. Advanced techniques like fluorescence-activated cell sorting (FACS) or microfluidics are employed to isolate individual microbial and host cells, ensuring minimal contamination and high viability.
Step 2: Nucleic Acid Extraction and Amplification
DNA and/or RNA are extracted from the isolated single cells. Low-input nucleic acid amplification methods are then applied to ensure sufficient material for high-throughput sequencing without introducing significant bias.
Step 3: Library Preparation and Sequencing
Prepared nucleic acid samples are converted into sequencing libraries. Our platforms utilize advanced next-generation sequencing (NGS) technologies, such as 16S rRNA gene sequencing (for bacterial identification) or shotgun metagenomic sequencing (for broader microbial and functional profiling), complemented by host single-cell RNA sequencing to profile host-microbe interactions.
Step 4: Bioinformatics Analysis and Data Interpretation
Raw sequencing data undergoes rigorous quality control, alignment, and bioinformatics analysis. This includes taxonomic classification, microbial abundance profiling, functional annotation (e.g., metabolic pathways, virulence factors), and integration with host single-cell data to understand cell-type-specific microbial associations. Advanced computational tools are used to identify microbial-host crosstalk and potential biomarkers.
Step 5: Quality Assurance and Reporting
Throughout the process, stringent quality control measures are implemented to ensure the reliability and accuracy of results. This includes monitoring library quality, sequencing depth, and reproducibility. A comprehensive report detailing methodologies, raw data, processed data, and interpretive analysis is provided.
Our Service Highlights
-
Unparalleled Resolution: Precisely identifies and characterizes individual microbial cells within the heterogeneous tumor microenvironment.
-
Revealed Interactions: Unveils subtle yet critical host-microbe interactions, often obscured by bulk sequencing methods.
-
Comprehensive Understanding: Provides insights into microbial diversity, functional potential, and spatial distribution.
-
Empowered Research: Facilitates the discovery of novel therapeutic targets and prediction of patient response to therapies.
-
Accelerated Development: Links specific microbes to cellular states and therapeutic outcomes, significantly accelerating cancer research and drug development.
FAQs
Q1: What types of samples are suitable for intratumoral microbiota single-cell analysis?
A1: We primarily recommend fresh or frozen tumor tissue samples to ensure optimal cell viability and microbial integrity. However, we can discuss the suitability of other sample types based on your specific research needs. Feel free to reach out for a detailed consultation on sample requirements.
Q2: How does single-cell analysis differ from bulk microbial sequencing, and why is it beneficial?
A2: Bulk sequencing provides an average microbial profile across an entire sample, masking cellular heterogeneity. Our single-cell analysis, conversely, allows for the characterization of individual microbial cells and their association with specific host cell types, revealing fine-grained interactions and rare microbial populations crucial for understanding tumor biology and therapeutic response. This deeper insight can lead to more targeted interventions.
Q3: What kind of data will I receive from the intratumoral microbiota single-cell analysis service?
A3: You will receive comprehensive bioinformatics reports, including taxonomic classification of identified microbes, their relative abundances, functional pathway analysis, and integrated analyses with host single-cell data. All data will be presented in a clear, interpretable format, with raw and processed data files available for your further investigation. We also offer interpretive sessions to walk you through the findings.
Q4: Can this service help in identifying novel therapeutic targets or biomarkers?
A4: Absolutely. By providing high-resolution data on microbial composition, function, and host interactions within the tumor, our service is designed to uncover novel microbial biomarkers for diagnosis, prognosis, and therapeutic response prediction. It also aids in identifying potential microbial targets for precision oncology strategies. We encourage you to discuss your specific target identification goals with our scientific team.
Q5: How does Creative Biolabs ensure the quality and accuracy of the single-cell analysis results?
A5: We employ stringent quality control steps at every stage, from sample processing and nucleic acid amplification to library preparation and sequencing. Our advanced bioinformatics pipelines are regularly validated, and our expert team performs thorough data interpretation, ensuring robust and reproducible results. We are committed to delivering reliable and accurate scientific findings to advance your research.
Why Choose Creative Biolabs
Creative Biolabs stands at the forefront of single-cell analysis, offering cutting-edge technology, unparalleled expertise, and a commitment to delivering high-quality, actionable data. Our integrated platform combines advanced microbial and host single-cell sequencing capabilities with sophisticated bioinformatics, ensuring a holistic understanding of the tumor ecosystem. We are dedicated to providing customized solutions that meet the unique needs of each research project, backed by rigorous quality control and experienced scientific support. We encourage you to contact us to work together for further advancement.
Reference
-
Lombardo, Claudia et al. "Intratumoral Microbiota: Insights from Anatomical, Molecular, and Clinical Perspectives." Journal of personalized medicine vol. 14,11 1083. 31 Oct. 2024. Distributed under Open Access License CC BY 4.0, without modification. DOI: https://doi.org/10.3390/jpm14111083


 Fig.1 Intratumoral microbes can promote metastasis formation in cancer cells.1
Fig.1 Intratumoral microbes can promote metastasis formation in cancer cells.1
