Next-generation Sequencing (NGS)-based Tumor Profiling
Overview
Next-generation sequencing (NGS) is one of the most advanced technologies applied to decipher molecular alterations in tumors and enables scientists to rapidly identify numerous mutations in patient tumors. It is an accurate and precise technology that provides comprehensive mutational profiling using as low as 10 ng DNA extracted from small biopsy specimens. Based on an NGS technology platform, Creative Biolabs offers a tumor profiling service for tumor biomarker discovery and development. We utilize our proprietary technology, extensive experience, and expertise to offer solutions to biotech and Pharma partners.
NGS-based Tumor Profiling Approaches
For precisely tailoring available targeted therapies to each individual or a subset of cancer patients, NGS has been utilized as a promising diagnostic tool with its advantages of accuracy, sensitivity, and high throughput. Current NGS approaches also differ based on the extent of target enrichment and sequencing involved, with the 3 major types being whole genome sequencing (WGS), whole-exome sequencing (WES), and targeted gene panels. Today, we offer a wide selection of NGS-based Tumor Profiling services that provide essential prognostic and therapeutic information for both hematologic and solid tumor diseases. Our solutions enable high-sensitivity detection of somatic single nucleotide variants (SNVs), indels, copy number variations (CNVs), and translocations in a single assay, supporting low-input DNA and formalin-fixed paraffin-embedded (FFPE) samples.
Whole Genome Sequencing
Whole genome sequencing (WGS) is able to detect genetic alterations in the entire genome. It provides a deep insight into the DNA sequence of human genomes, with data analysis at the individual or population level. SNP/INDEL/CNV/SV and other genome variants can be fully analyzed.
Whole Exome Sequencing
Whole exome sequencing (WES) is able to detect genetic alterations in the exon regions. By focusing on the entire protein-coding regions of the genome, WES guides you to reach a diagnosis sooner, avoiding extensive, costly and lengthy serial testing.
Targeted Sequencing
Targeted sequencing is able to sequence key genes or regions of interest to high depth, allowing identification of rare variants. This focused approach yields deeper sequencing of target regions with fewer total reads compared to WGS, reducing the amount of sequencing needed and the time required for analysis.
Comparison of NGS Approaches
The choice between NGS approaches depends on several factors, including the final application of tumor testing (clinical vs research), the results required, technical efficiency, and cost. You can choose the sequencing service you need according to your needs. Our experienced team of experts is involved throughout the process to provide you with relevant consulting services and sequencing advice.
Key Features & Advantages
Creative Biolabs is the original, most experienced comprehensive molecular profiling laboratory, and we have optimized our systems to provide industry-leading reports, service and turnaround time.
Our NGS-based tumor profiling service provides a comprehensive method for assessing the majority of genes associated with tumors, including lung, colon, breast, melanoma, gastric, and ovarian cancers.
Our team of talented and experienced computational biologists use internally developed tools and pipelines to help you better understand your data and the underlying biology of your experiments.
Service Workflow
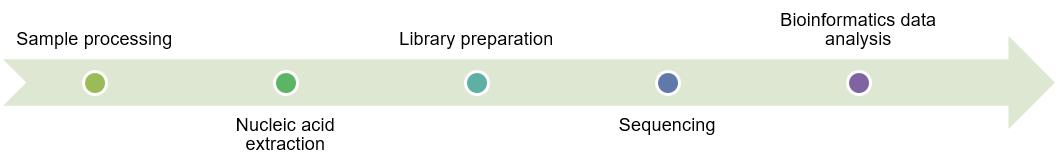
The basic methodology of NGS includes nucleic acid extraction and sample processing, library preparation, amplification, sequencing, and bioinformatics data analysis and interpretation.
Sample Requirements
Sample Source: Total DNA from cells, blood, saliva, tissue, fresh-frozen tissue biopsy, FFPE tissue, cancer liquid biopsies, etc.
|
NGS Approach
|
Sample Types
|
Recommended Quantity
|
Sample Purity
|
Minimum Amount
|
|
WGS
|
Genomic DNA (gDNA) from blood, fresh-frozen biopsy
|
>1.5µg, >20 ng/µL
|
(OD260/280):1.8-2.0
|
500 ng
|
|
WES
|
gDNA from blood, fresh-frozen biopsy
|
>1.5µg, >20 ng/µL
|
(OD260/280):1.8-2.0
|
500 ng
|
|
Targeted Sequencing
|
gDNA and/or RNA from blood, fresh-frozen biopsy; DNA and RNA from FFPE
|
>1µg, >20 ng/µL
|
(OD260/280):1.8-2.0
|
100 ng
|
Any specific requirements please contact us.
Published Data
Data 1: Whole-genome sequencing and alignment of colorectal cancer sample.
Sample: Genomic DNA (gDNA) from the fresh-frozen biopsy.
Approach: WGS
 Fig.2 Oncoprint representation of recurrently mutated genes. (Ishaque, et al., 2018)
Fig.2 Oncoprint representation of recurrently mutated genes. (Ishaque, et al., 2018)
Data 2: Whole-exome sequencing and alignment of small cell lung cancer.
Sample: Genomic DNA (gDNA) from the fresh-frozen biopsy.
Approach: WES
 Fig.3 Mutational spectrum of small cell lung cancer. (Zhou, et al., 2021)
Fig.3 Mutational spectrum of small cell lung cancer. (Zhou, et al., 2021)
Contact Us
Discuss Your Project Requirements
Whether you want to know more information about our service or would like expert support with project design, we can deliver.
Feel free to contact us.
References
-
Ishaque, Naveed, et al. "Whole genome sequencing puts forward hypotheses on metastasis evolution and therapy in colorectal cancer." Nature communications 9.1 (2018): 4782.
-
Zhou, Huaqiang, et al. "Multi-region exome sequencing reveals the intratumoral heterogeneity of surgically resected small cell lung cancer." Nature communications 12.1 (2021): 5431.
-
Distributed under Open Access license CC BY 4.0, without modification.
For Research Use Only | Not For Clinical Use






 Fig.2 Oncoprint representation of recurrently mutated genes. (Ishaque, et al., 2018)
Fig.2 Oncoprint representation of recurrently mutated genes. (Ishaque, et al., 2018)
 Fig.3 Mutational spectrum of small cell lung cancer. (Zhou, et al., 2021)
Fig.3 Mutational spectrum of small cell lung cancer. (Zhou, et al., 2021)

 Download our brochure
Download our brochure