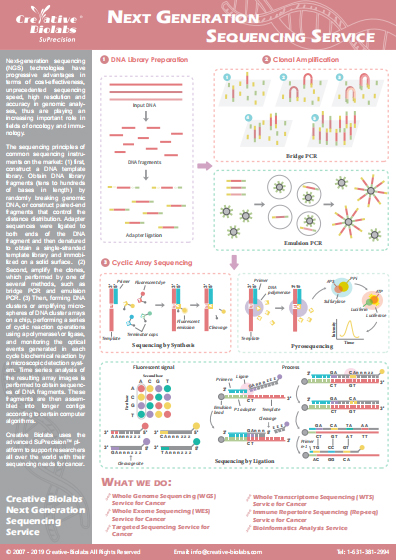
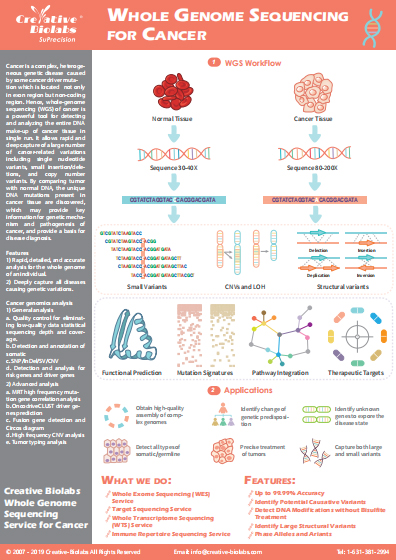
Three-dimensional (3D) Genomic Service
The three-dimensional (3D) configuration of the genome is complex and dynamic. To understand the workings of the genome, we must study the chromosome structure. With years of experience in applying chromosome conformation capture technologies (C-technologies), Creative Biolabs has gained significant knowledge in 3D genomics. We are more than happy to provide 3D genomics service using high-throughput chromosome conformation capture (Hi-C) technology.
The Introduction of 3D Genome
The nucleus of a eukaryote is a complex 3D environment. Eukaryotic genome function depends not only on the linear arrangement of regulatory sequence elements but also on their spatial organization for effective control of gene expression. The interactions between regulatory elements and gene promoters are essential for organismal development and response to environmental stimuli. Various technologies have demonstrated that the genome is organized in a non-random fashion into higher-order chromatin domains.
C-technologies
Analysis of the role of chromatin 3D organization in gene expression is progressing rapidly, largely due to the technological advances, such as chromosome conformation capture (3C), circular chromosome conformation capture (4C), carbon copy chromosome conformation capture (5C) and Hi-C methods. These technologies reveal how the organization of the genome is interconnected with nuclear architecture and can vary between cell types and during cell differentiation and development.
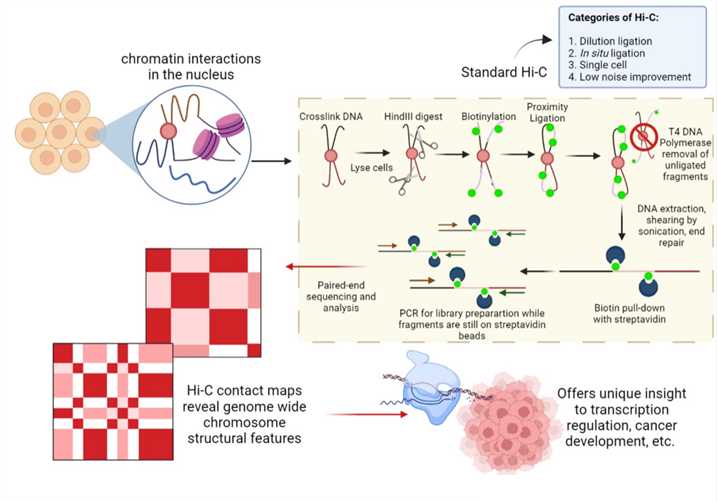 Fig 1. Schematic diagram of Hi-C. Distributed under CC BY-SA 4.0, from Wiki,
without modification.
Fig 1. Schematic diagram of Hi-C. Distributed under CC BY-SA 4.0, from Wiki,
without modification.
- 3C (one vs one method)
This method involves formaldehyde cross-linking of cells followed by chromatin isolation and digestion with a restriction enzyme. Specific enzymes are used to free a known or predicted DNA-DNA interaction mediated by a protein complex. These freed fragments are then ligated into rings and the crosslinks are reversed. Real-time PCR is then used to determine the ligation product abundance in the purified DNA. The abundance directly correlates to the interaction frequency of the two ligated regions. This approach allows any candidate chimeric product generated from the crosslinking step to be quantified.
- 4C (one vs all method)
In this method, libraries of chimeric DNA products are digested with a second restriction enzyme and further re-ligated together to decrease their molecular size. Using circular PCR, all chimeric products ligated with a desired specific restriction fragment are amplified and sequenced, generating an interaction profile measuring all DNA fragments interacting with the desired fragment. This approach allows the identification of previously unknown DNA regions that interact with a locus of interest, which makes it especially suited to discover novel interactions with a specific region.
- 5C (many vs all method)
5C uses special primers with a universal sequence in the tail that allows amplification of all ligation products. This technique generates a library of any ligation products from DNA regions that associate with the target loci, which are then analyzed by next-generation sequencing (NGS). 5C is ideal to find great detail about all the interactions in a given region. However, since each 5C primer must be designed individually, 5C is best suited to particular regions but not truly genome-wide.
- Hi-C (all vs all method)
Template generation in Hi-C is similar to that in 3C, but there is a slightly modified ligation step. Following restriction digestion of crosslinked nuclear DNA, the DNA overhangs are filled in with dNTPs, one of which is biotinylated. The resulting DNA blunt ends are subsequently ligated. After that, DNA is purified and sequenced using paired-end sequencing, which produces a genome-wide interaction map of ‘all vs all’ restriction fragments.
3D Genomics Service at Creative Biolabs
With Ph.D. level scientists in applying Hi-C technology, 3D position of chromatin in the nucleus can be comprehensively interrogated. Based on our advanced SuPrecision™ Platform, we provide important insights into underlying mechanisms that may be responsible for various human diseases including cancer.
Hi-C provides a powerful tool to better characterize 3D chromatin organization. Its applications include but are not limited to:
- Studying human genetic disease
- Uncovering the impact of cancer risk-associated single nucleotide polymorphisms (SNPs)
- Classification of cancer subtypes
- Alleviating cancer progression by inhibiting certain chromatin loop formations
Equipped with world-leading technology platforms and professional scientific staff, Creative Biolabs provides high-quality 3D genomics service using Hi-C technology. Please contact us for more information and a detailed quote.
Resources
Infographics
Podcast
- Whole Genome Sequencing (WGS)
- Whole Exome Sequencing (WES)
- Targeted Sequencing
- Whole Transcriptome Sequencing (WTS)
- Immune Repertoire Sequencing (Rep-Seq)
- Epigenomics
- Metagenomic Sequencing
- ctDNA Sequencing
- Single Cell Sequencing
- Spatial Transcriptome Sequencing