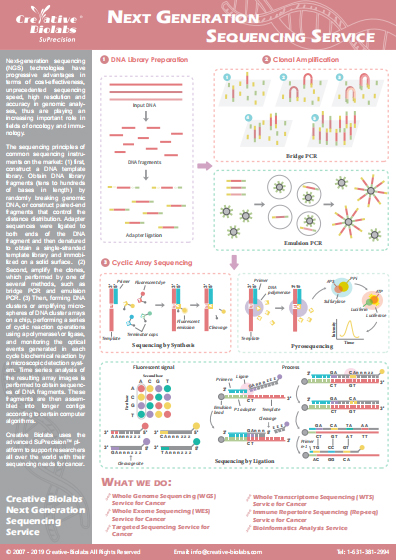
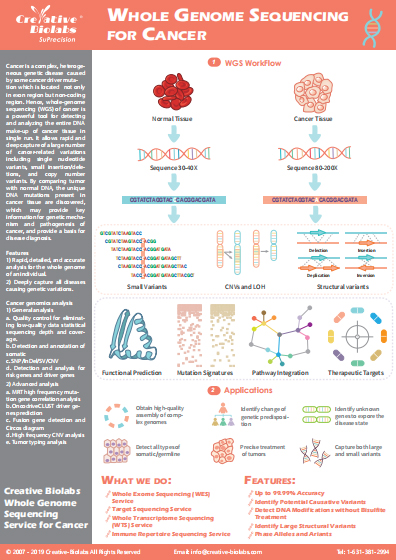
Metagenomic Sequencing Services for Cancer
Metagenomics is an equitable way of identifying microbiota present in the tissue specimens of cancer patients. In cancer research, this approach has revolutionized the way to identify, analyze and target microbial diversity. Based on our advanced SuPrecision™ Platform, Creative Biolabs is pleased to share our cutting-edge technology and extensive expertise in metagenomics sequencing for cancer-associated microbiota to facilitate our clients’ project development.
Metagenomic Studies in Cancer
Metagenomics refers to all the environmental communities and comprises the genomes of hundreds or more organisms from all three domains of life i.e. archaea, bacteria and eukarya, as well as viruses. Considering the fact that thousands of microbial species comprise the normal human microbiome, it is likely that the microbe communities can substantially influence normal physiology, as well as the major contributors to and response to diseases, including cancer. Statistically, about 16% of cancers are associated with microbial infection. Thus, studying the metagenomics of patient-derived tumor tissues will provide a better understanding of the contributory role of the microbiome in therapeutic responses to-associated cancer. As a horizon in cancer research, metagenomics has broadened the scope of targeting microbes responsible for various types of cancers. Currently, metagenomics has been paid more and more attention in pharmaceutical and biotechnological industries for the discovery of novel drugs.
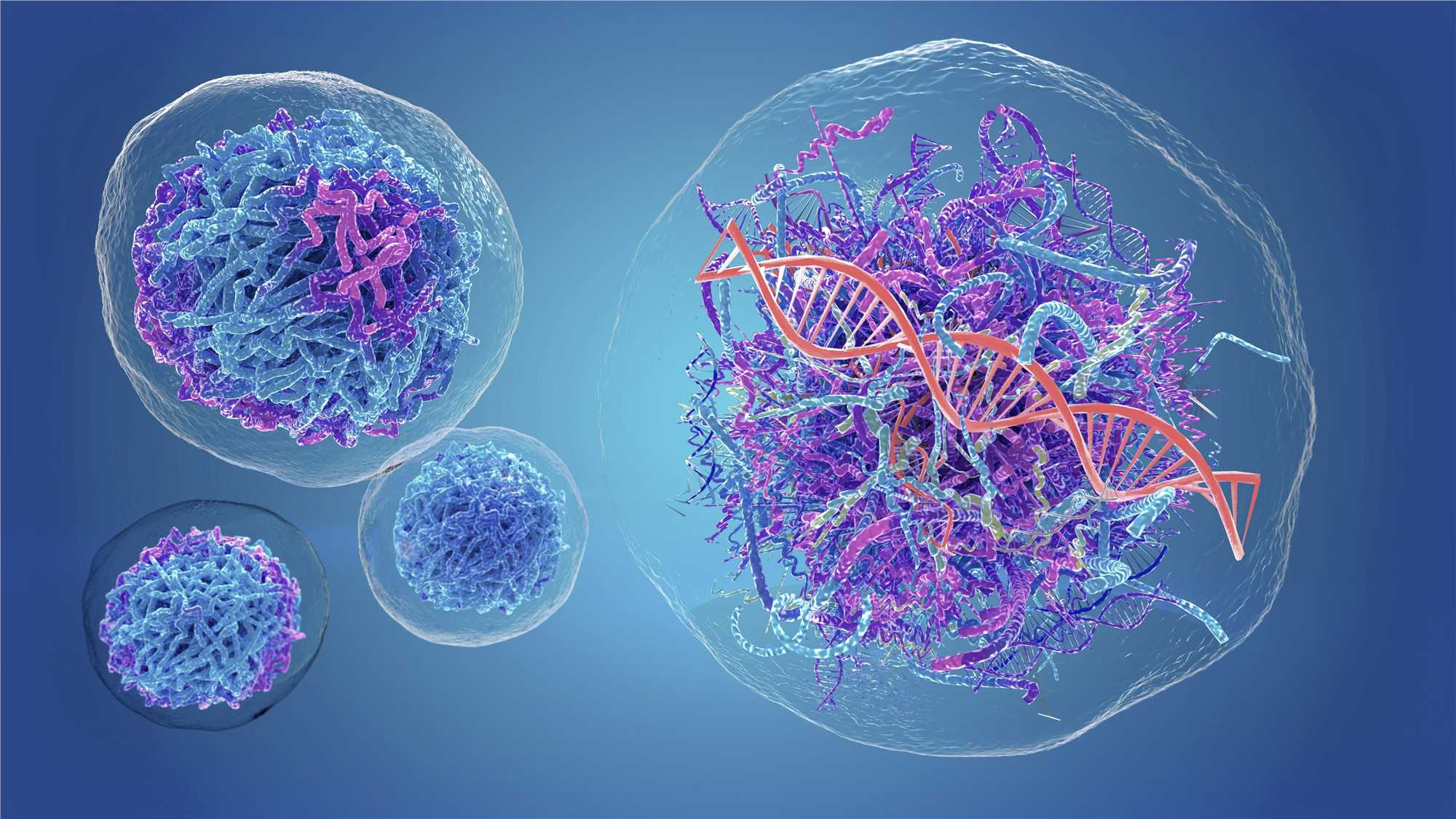
Cancer-associated Microbiota
- Colorectal cancer (CRC)-associated microbiota
Among the top three commonest cancers globally, CRC is a complex disease influenced by genetic and environmental factors. Acquired evidence suggests that the human gut microbiome is related to the development of colorectal cancer. Gut microbiota (> 1013 microorganisms) colonizes the human gastrointestinal tract with a role to maintain gut integrity, metabolism, immunity, and protect against pathogens. The gut microbiota consists of commensal organisms that include bacteria, archaea, eukarya, and viruses.
- Prostate cancer-associated microbiota
Prostate cancer is the most common type of noncutaneous cancer diagnosed in men and the second leading cause of cancer-related death. Metagenomic studies have shown that the most abundant genera in the sample of patients with prostate cancer are Escherichia, Propionibacterium, Acinetobacter and Pseudomonas, although the exact quantitative compositions varied. Moreover, four viruses were identified in the metagenome, all of which belong to dsDNA viruses.
- Breast Cancer-associated microbiota
Though age, diet and genetic predisposition are established risk factors, the majority of breast cancers have unknown etiology. In a qualitative analysis of microbiota DNA of breast, scientists have found that the bacterium Methylobacterium radiotolerans is relatively enriched in tumor tissue, while the bacterium Sphingomonas yanoikuyae is relatively enriched in paired normal tissue.
- Gastric cancer-associated microbiota
Gastric cancer is the third leading cause of cancer-related deaths worldwide, with an estimated 1 million new cases recorded yearly. Dysbiosis of gastric microbiota, such as Helicobacter pylori, plays a significant role in the pathogenesis and progression of gastric cancer. Low pH in the gastric secretions favors the growth of bacteria. Studies on animal models also confirmed the role of gastric microbiota in the development of gastric cancer. The predominant microbial population in the gastric cancer patients have been found to be Veillonella, Haemophilus, Streptococcus, Lactobacillus, Prevotella and Neisseria spp.
- Oral carcinoma-associated microbiota
The microbiota residing in the oral cavity are often detrimental and give rise to different oral diseases including oral cancers. Previous studies found Prevotella melaninogenica and Capnocytophaga gingivalis and Streptococcus mitis in the oral squamous cell carcinoma (OSCC), and suggested that salivary microbiotas can be used as the diagnostic marker for oral cancers.
Metagenomics Sequencing Service at Creative Biolabs
Creative Biolabs is fully competent and dedicated to serving as your one-stop-shop for metagenomics sequencing of cancer-associated-bacteria or virus. The general process includes 1) sample processing, 2) prevention of contamination and negative controls, 3) library construction and sequencing, 4) filtering human DNA and mapping to microbial genomes, 5) taxonomic analysis, 6) functional analysis. Our scientists are skilled in rapidly acquiring data on the metagenomic landscapes of different cancers.
Key Advantages
- Various types of cancer sample are available
- Sophisticated bioinformatic tools
- Powered by a proven, first-in-class technology
- Best after-sale service
With years of experience in metagenomics sequencing and bioinformatics analysis, Creative Biolabs is dedicated to providing high-quality metagenomics sequencing service for cancer. Valuable data will be presented to customers, which will expand our understanding on the oncogenic mechanisms and offer new opportunities to develop new applications for cancer diagnosis and treatment. Please contact us for more information and a detailed quote.
Resources
Infographics
Podcast
- Whole Genome Sequencing (WGS)
- Whole Exome Sequencing (WES)
- Targeted Sequencing
- Whole Transcriptome Sequencing (WTS)
- Immune Repertoire Sequencing (Rep-Seq)
- Epigenomics
- 3D Genomics
- ctDNA Sequencing
- Single Cell Sequencing
- Spatial Transcriptome Sequencing