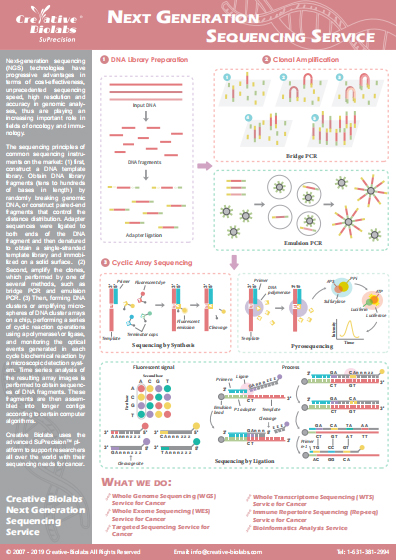
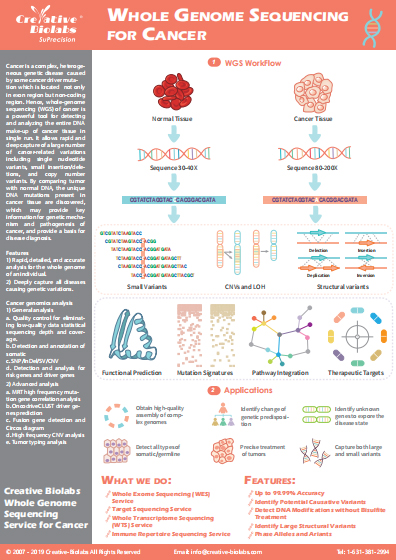
Single-Cell DNA Methylation Sequencing Service
Single-cell DNA methylation sequencing enables us to understand the heterogeneity of genome-wide 5-methylcytosine (5mC) patterns, providing a comprehensive and powerful platform for tumor diagnosis and precision therapy. As a leader in NGS-based cancer research, Creative Biolabs offers unrivaled single-cell whole-genome bisulfite sequencing services based on our advanced and powerful sequencing platforms.
Introduction of DNA Methylation
DNA methylation is an epigenetic modification that occurs by the addition of methyl (CH3) groups to the DNA molecule. It plays a key role in the regulation of gene expression without changing the gene sequence. In mammals, DNA methylation is associated with many key biological processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis. Furthermore, DNA methylation is also associated with tumorigenesis. Increased evidence has reported aberrant DNA methylation is involved in the development and progression of human cancers. Tumor cells may display hypomethylation, leading to an increased oncogene activity which may be suppressed in normal cells. While tumor suppressor genes display hypermethylation which renders them inactive. Changes in DNA methylation in cancer can be considered as a promising target for the development of powerful diagnostic, prognostic, and predictive biomarkers.
Single-cell Whole Genome Bisulfite Sequencing
The identical individual organisms in almost all of the different types of cells have almost exactly the identical sequence of genome DNA. These DNA sequences may have different methylation modifications, thus making each type of cell, even every cell has its unique characteristics of gene expression, furthermore resulted in heterogeneity between different types of cells or between different cells of the same cell type. Single-cell whole-genome bisulfite sequencing combines bisulfite treatment and high-throughput sequencing technologies to detect genome-wide methylation at a single-cell level, making it possible to explore the epigenetic information of highly heterogeneous cells.
Workflow
 Fig.2 Workflow diagram.
Fig.2 Workflow diagram.
Bioinformatics
| General Analysis | Senior Analysis |
|---|---|
|
|
Our Advantages
- Well-established single-cell whole-genome bisulfite sequencing technology
- An experienced expert team providing one-stop services from preparation of cell suspension to high-quality data delivery
- Personalized and customized data analysis services
Sample Requirement
- Sample type: cell lines, primary cells, fresh tissue, cryopreserved cells
- Species: Humans, mice, and other species need to be evaluated
- Storage and transportation: Stored in liquid nitrogen, transported on dry ice
If you are interested in our services, please feel free to contact us for more details.