Why iPSC Characterization Matters
iPSCs are inherently sensitive, highly plastic, and prone to genomic or phenotypic drift during culture. Without systematic characterization, even subtle abnormalities can compromise the reproducibility of differentiation, distort disease phenotypes, or generate misleading conclusions. Robust evaluation therefore becomes essential for safeguarding both the scientific integrity of your project and the translational potential of your iPSC lines.
High-quality characterization matters for several critical reasons:
- Confirming True Pluripotency: Marker expression (OCT4, SOX2, NANOG), teratoma assays, EB formation, and qPCR validation ensure that the cells retain the molecular signatures necessary for consistent performance across studies.
- Ensuring Genomic Stability for Downstream Reliability: Karyotype analysis ensures researchers work with genetically stable lines suitable for disease modeling, drug screening, and translational research.
- Supporting Functional Competence: Beyond molecular markers, functional readouts—such as electrophysiological activity from MEA assays—demonstrate whether differentiated cells perform as expected. For applications involving neurons or cardiomyocytes, functional validation is indispensable for assessing maturity, excitability, and responsiveness to compounds.
Our iPSC Characterization Services
| Services | Descriptions |
|---|---|
| Morphology of iPSC |
Cell morphology provides the earliest visual indicator of stemness and culture quality. Our specialists assess:
|
| Teratoma Formation Assays for iPSC |
Teratoma assays represent the gold standard for pluripotency evaluation in vivo. We offer:
|
| Embryoid Body (EB) Formation and Characterization for iPSC |
EB formation replicates spontaneous differentiation in a controlled, 3D culture environment. Our workflow includes:
|
| Karyotype Analysis for iPSC |
Genomic stability is essential for safety, differentiation reliability, reproducibility, and regulatory acceptance. We provide:
|
| Gender Determination of iPSC |
Sex chromosome identity may influence differentiation efficiency and disease-model relevance. Creative Biolabs offers:
|
| Electrophysiological Characterization via Multi-electrode Array (MEA) for iPSC |
MEA analysis provides functional readouts particularly important for iPSC-derived neurons and cardiomyocytes. Our capabilities include:
|
| qPCR Analysis for Pluripotency Markers for iPSC |
Gene expression analysis ensures that your iPSC lines maintain high pluripotency. We evaluate key markers such as:
|
| Pluripotency Marker Assays for iPSC |
Using immunofluorescence and flow cytometry, we evaluate surface and intracellular markers including:
|
Technology Platforms Supporting Our Characterization Assays
We leverage a broad suite of high-precision instruments:
- High-content imaging systems for morphology and immunostaining
- qPCR and digital PCR platforms
- Flow cytometry systems
- G-banding cytogenetics platforms
- MEA electrophysiology arrays (high-throughput)
- Automated histology systems
- Spectral imaging platforms for marker quantification
- Genomic QC systems (optional add-on: CNV detection, STR profiling)
This robust infrastructure ensures high data fidelity and rapid delivery.
How We Solve The Key Challenges in iPSC Characterization
Characterizing iPSCs is technically demanding due to their plasticity and sensitivity. Creative Biolabs helps clients overcome typical bottlenecks.
-
Culture Variability Between Laboratories - Even minor differences in media, feeder systems, or passaging protocols may alter pluripotency.
Our solution: standardized SOPs, harmonized culture conditions, and side-by-side comparisons with reference cell lines. -
Genetic Instability During Extended Expansion - Long-term culture increases the risk of chromosomal abnormalities.
Our solution: periodic karyotyping and optional rapid genomic integrity screening. -
Loss of Pluripotency - iPSCs may gradually lose stemness or partially differentiate.
Our solution: multi-parameter evaluation including morphology, immunostaining, qPCR, and EB assays to detect early deviations. -
Inefficient or Inconsistent Differentiation - Inadequate initial quality leads directly to poor differentiation performance.
Our solution: teratoma and EB formation assays that confirm tri-lineage potential prior to downstream applications. -
Lack of Functional Validation - Some projects require proof that differentiated cells behave as expected.
Our solution: high-resolution MEA electrophysiology for neurons/cardiomyocytes, paired with cell-type-specific markers.
Wide Applications of iPSC Characterization
High-quality iPSC characterization is not merely a quality-control requirement—it is the engine that drives the success of a broad spectrum of scientific, preclinical, and industrial programs. Robust characterization data ensures that the iPSC lines they rely on are genetically stable, phenotypically consistent, fully pluripotent, and capable of differentiating into functional, lineage-specific cells.
Below, we demonstrate how high-standard iPSC characterization can empower key application scenarios, according to industry needs and research processes.
| Applications | Descriptions |
|---|---|
| Disease Modeling | Accurately representing patient-specific phenotypes (e.g., neurodegenerative, cardiovascular, metabolic disorders). |
| High-Throughput Drug Screening | Ensures differentiated cells respond reproducibly to candidate compounds. |
| Toxicity and Safety Evaluation | iPSC-derived hepatocytes, cardiomyocytes, and neurons form robust in vitro toxicity platforms. |
| Cell Therapy Development | Characterization ensures genomic stability, lineage identity, and phenotype fidelity before moving to translational stages. |
| iPSC Banking & Reference Line Development | Standardized QC ensures long-term reliability of master and working cell banks. |
| Personalized Medicine | Sex verification and genomic stability are essential for individualized disease models. |
Published Data
The researchers used the popular 3D floating culture method to generate retinal organoids from stem cells. This method starts with either small clumps of stem cells generated from larger clones (clumps protocol, CP) or with an aggregation of single cells (single cells protocol, SCP). Using histological analysis and gene-expression comparison, they found a retention of the pluripotency capacity on embryoid bodies generated through the SCP compared to the CP.
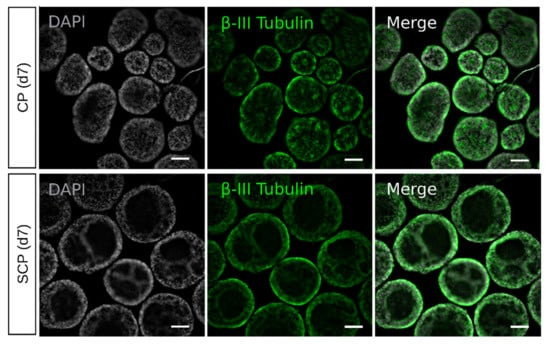 Fig. 1 Confocal imaging of EBs generated using the CP and the SCP at day 7.1,3
Fig. 1 Confocal imaging of EBs generated using the CP and the SCP at day 7.1,3
The researchers discussed the different types of MEAs used for in vivo and in vitro recordings. They summarized 2D hPSC-derived neural cultures on MEA, their strengths, weaknesses, and what kind of information they can provide us on the physiology and pathology of neuronal networks. As organoids and other hPSC-derived 3D models have already been in use for several years, they took a similar look into their MEA data, their ins, outs, and how they can describe the structure and function of the human brain.
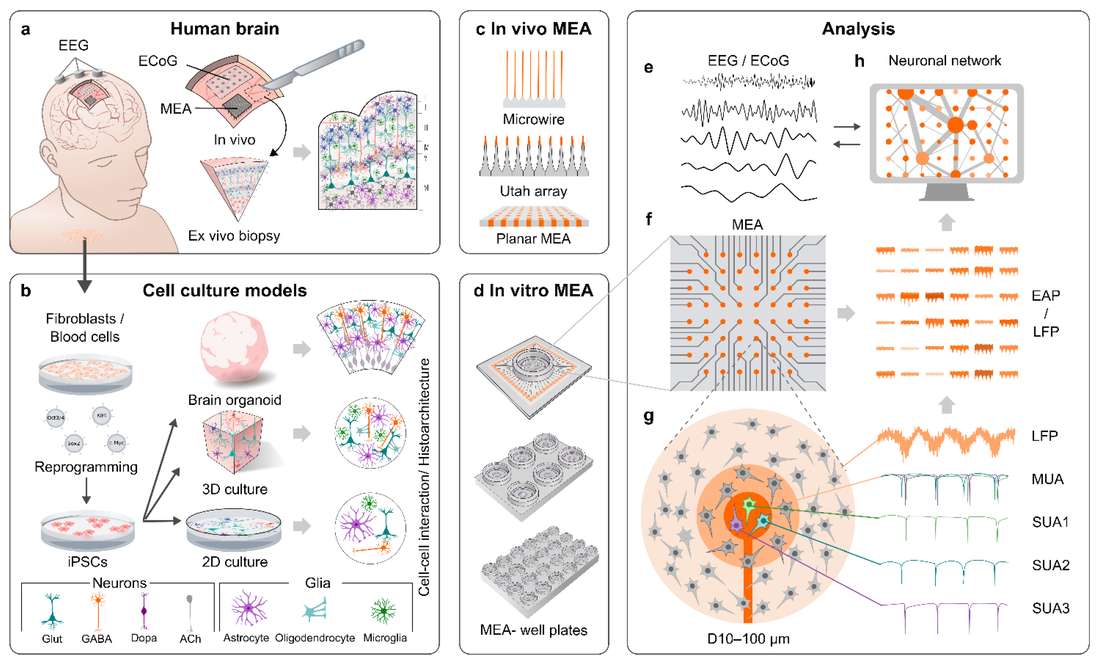 Fig. 2 MEA recordings of the human brain and corresponding hPSC-derived models.2,3
Fig. 2 MEA recordings of the human brain and corresponding hPSC-derived models.2,3
What Our Clients Say
"We sent Creative Biolabs several newly reprogrammed iPSC lines. Their morphology assessment, pluripotency staining, and qPCR analysis were extremely thorough. The report not only confirmed pluripotency but also highlighted early differentiation tendencies we hadn't noticed. This level of detail saved us months of downstream troubleshooting."
— Principal Investigator, Stem Cell Biology Lab
"Our team needed stable and well-characterized iPSC lines for a neurological screening platform. Creative Biolabs provided prompt karyotype results, clear EB differentiation data, and a clean QC package that our automation team could rely on. It made our transition to screening much smoother."
— Director of Drug Discovery, Mid-size Biopharma
"We worked with their MEA team to validate our iPSC-derived neurons. The field potential recordings, spike analysis, and network activity metrics were highly detailed. Their scientific support helped us interpret the patterns and prepare figures for a manuscript."
— Senior Scientist, Neuroscience Program
"Some of our disease models are sex-specific. Creative Biolabs helped us validate XX/XY identity and confirm the authenticity of each iPSC line. Their responsiveness and scientific clarity make them easy to work with."
— Group Leader, Personalized Medicine Research Center
FAQs
Q: What level of characterization is recommended before starting an iPSC differentiation project?
A: A comprehensive panel including morphology assessment, pluripotency marker profiling, qPCR validation, and karyotype analysis is typically sufficient. For projects involving functional cell types such as neurons or cardiomyocytes, EB formation assays and optional MEA evaluations are strongly recommended.
Q: Can Creative Biolabs work with iPSC lines generated from Sendai, episomal, mRNA, or viral reprogramming?
Q: How many cells should we prepare for a full characterization package?
Q: Can results from multiple characterization assays be combined into a single integrated report?
Q: What if our iPSC line has minor chromosomal abnormalities? Can we still proceed?
Q: What quality controls are performed before you begin characterization?
Get Started with Confidence














1. Contact Us
via the Inquiry Form or Email
2. Define Your Needs
Cell Type, Function, Quantity, Modifications
3. Kickstart the Project
Our Expert Team Guiding Every Step
High-quality data is the foundation of reliable stem cell research. Creative Biolabs combines industry-leading technology platforms with deep scientific expertise to deliver comprehensive, reproducible, and actionable iPSC characterization.
Contact us today to discuss your project or request a customized quote.
References
- Heredero Berzal, Andrea, et al. "The Analysis of Embryoid Body Formation and Its Role in Retinal Organoid Development." International Journal of Molecular Sciences 25.3 (2024): 1444. https://doi.org/10.3390/ijms25031444
- Pelkonen, Anssi, et al. "Functional characterization of human pluripotent stem cell-derived models of the brain with microelectrode arrays." Cells 11.1 (2021): 106. https://doi.org/10.3390/cells11010106
- Distributed under Open Access license CC BY 4.0, without modification.
Created November 2025