SuPrecision™ Pipeline for Immune Repertoire Analysis
Creative Biolabs is a long-term expert in the field of immunology with extensive experience and advanced SuPrecision™ platform, which is mainly based on next-generation sequencing (NGS) technique. Our scientists have developed a unique bioinformatics pipeline for immune repertoire sequencing analysis. It can be an individual service and also can be provided for our customers cooperating with high-throughput sequencing services together. This method is not only suitable for the analysis of B cell receptor (BCR) sequencing data, but also T cell receptor (TCR) sequencing data.
The Requirement of Effective Bioinformatics Pipelines
High-throughput NGS technology is revolutionizing our ability to carry out large-scale BCR profiling studies. BCR repertoire sequencing data analysis is different from genomes sequencing and transcriptomes sequencing because BCR sequences are not encoded directly in the genome. While the V, D and J segments of the BCR can be traced back to segments encoded in the germline, the set of segments used by each receptor needs to be inferred, because it is coded in a highly repetitive region and currently cannot be sequenced directly. Furthermore, these segments can be significantly modified during the rearrangement process and through somatic hypermutation. Thus, there are no full-length templates for the sequencing reads alignment. As sequencing technologies continue to improve, these repertoire sequencing experiments are producing ever larger datasets. The researchers require specialized bioinformatics pipelines to analyze these data effectively. Although numerous methods and tools have been developed for the analysis, the results vary widely, and it is difficult to understand the result for non-professionals.
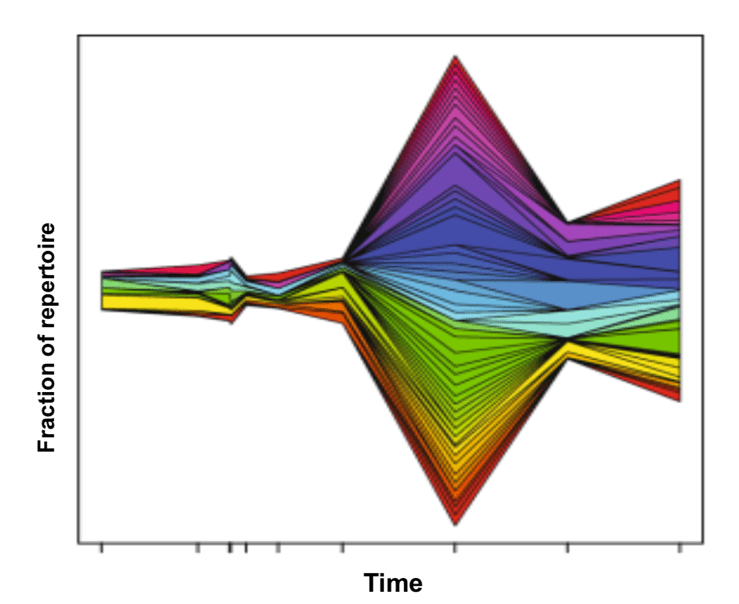 Fig.1 Overlap between time points, and the stream plot showing how clones expand and contract over time (Yaari & Kleinstein 2015).
Fig.1 Overlap between time points, and the stream plot showing how clones expand and contract over time (Yaari & Kleinstein 2015).
SuPrecision™ Pipeline for Immune Repertoire Analysis at Creative Biolabs
Creative Biolabs has developed a simple but useful pipeline for BCR repertoire sequencing analysis, starting from pre-processing, V(D)J germline segment assignment, and analysis of repertoire properties. Pre-processing includes quality control and read annotation and assembly of paired-end reads. Segment assignment includes methods for dynamic population structure, clonal grouping and B cell lineage trees. Repertoire analysis consists of diversity, somatic hypermutation analysis, stereotypic sequences and convergent evolution analysis. This approach is practical to solve bioinformatics problems in repertoires analysis.
Key Advantages of Immune Repertoire Analysis Pipeline Include but Are Not Limited to:
- Suitable for both BCR and TCR repertoire analysis
- Repertoire analysis at the single-cell level
- Identification of germline genes and alleles
- Highly professional Ph.D. level scientists
- Easy to understand for non-professionals
- Time-saving and price favorable
Creative Biolabs has accomplished over hundreds of sequencing projects and data analysis. Our scientists have accumulated extensive experience in BCR and TCR repertoire sequencing analysis. We are pleased to offer the best service with the most accurate results for our global customers.
Please contact us for more information and a detailed quote.
Reference
- Yaari, Gur, and Steven H. Kleinstein. "Practical guidelines for B-cell receptor repertoire sequencing analysis." Genome medicine 7 (2015): 1-14. Distributed under open access license CC BY 4.0, without modification.
Resources
Infographics
Podcast
- WGS Bioinformatic Analysis
- WES Bioinformatic Analysis
- Targeted Sequencing Bioinformatic Analysis
- WTS Bioinformatic Analysis