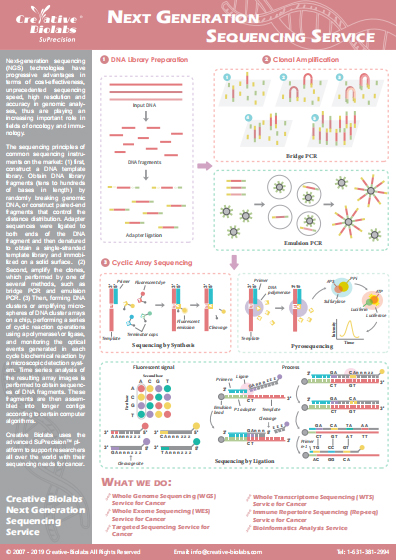
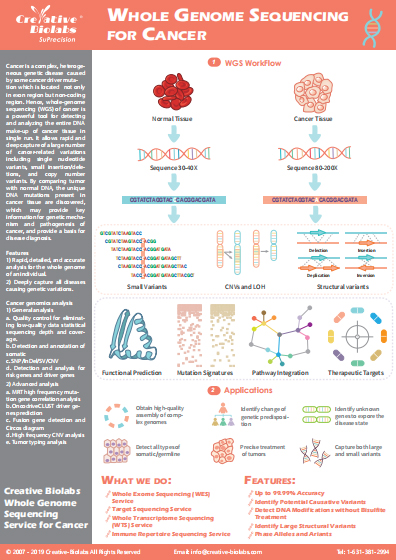
Immune Repertoire Germline Gene and Allele Identification Service
Next-generation sequencing (NGS) combined with bioinformatics is a powerful tool for analyzing the large number of sequences present in the immune repertoire. Creative Biolabs has developed a robust tool for predicting germline V/J genes and alleles through deep-sequencing data derived from B cell receptor (BCR) and T cell receptor (TCR) repertoires. We are proud to introduce this high-quality service to our clients all over the world. It can be an individual service, and it also can be provided for our clients cooperating with high-throughput sequencing services together.
More Germline Genes and Alleles Are Needed to Be Identified
The immune repertoire is composed of diverse BCRs and TCRs created by somatic recombination of many germline V (variable), D (diversity), J (joining), and C (constant) gene segments. In recent years, advances in NGS technology have enabled assessment of millions of BCRs or TCRs from a single sequencing assay. A large-scale study of the properties of TCRs and BCRs repertoires through NGS is providing excellent insights into the understanding of adaptive immune responses. Sequencing of the repertoire (Rep-seq) is being widely applied in academic and biomedical fields. V(D)J germline genes and alleles must be characterized comprehensively to facilitate repertoire analyses. The publicly available ImMunoGeneTics (IMGT) database only collects the genes of certain species. In conventional polymerase chain reaction (PCR)-based method, primers are designed based on human germline genes and are used to extract the counterparts of other species. Although this method has obtained high accuracy, it is suitable only for species that are extremely homologous to humans. So far, most species do not have well-characterized BCR/ TCR germline genes. Also, more alleles are required for both humans and other species, however, a validated method is lacking.
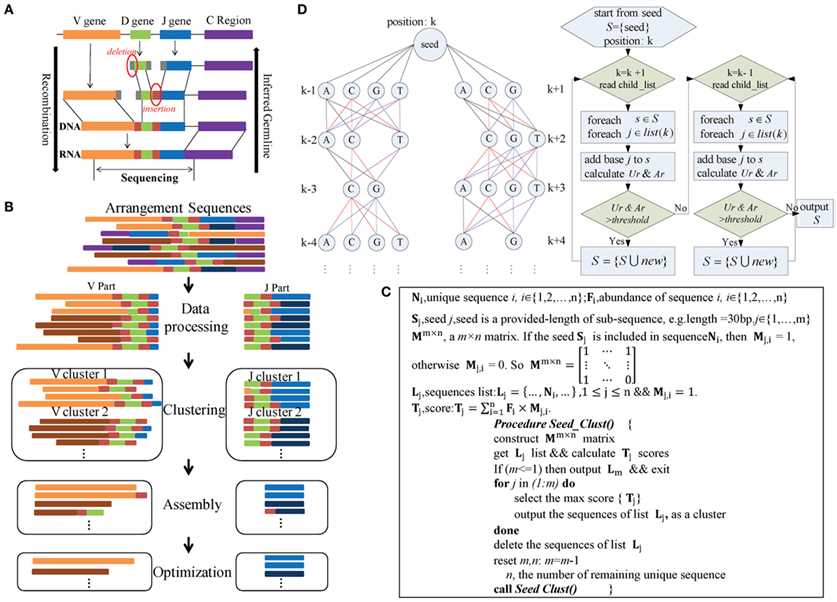 Fig. 1 Immune repertoire germline identification.1
Fig. 1 Immune repertoire germline identification.1
Germline Genes and Alleles Identification Service in Creative Biolabs
To resolve these issues, a tool to predict more and more novel germline genes and alleles is needed. Creative Biolabs has employed a novel tool to predict BCR/TCR germline genes and alleles using Rep-seq data without known germline sequences or assembled genome data. Without knowing V/J gene segments, primers designed at the C region are available for all species. Our method comprises four main steps: data processing, a clustering algorithm to classify sequences, assembly by a multiway tree structure and optimization. All highly accurate results we have obtained suggest this tool is stable with different species and long-sequence data.
Key Features and Advantages of Including but Not Limited to:
- Suitable for both human and other species
- No need of known germline sequences or assembled genome data
- High accuracy (>90%)
- Advanced SuPrecision™ platform
- Highly professional Ph.D. level scientists
As a long-term expert in the field of immunology, Creative Biolabs has accumulated extensive experiences. With the rapid accumulation of high-throughput sequence data for TCRs and BCRs repertoires, this powerful tool can be applied broadly for obtaining novel genes and a large number of novel alleles. We are confident in providing clients with the best service at the most competitive cost.
Please contact us for more information and a detailed quote.
Reference
- Zhang, Wei, et al. "IMPre: an accurate and efficient software for prediction of T-and B-cell receptor germline genes and alleles from rearranged repertoire data." Frontiers in immunology 7 (2016): 457. Distributed under open access license CC BY 4.0, without modification.
Resources
Infographics
Podcast
- TMB Analysis for Checkpoint Immunotherapy Response
- WES based Variant Analysis
- WES based Structural Variant Detection
- WES based CNV Detection
- WES based Tumor-Specific Neoantigen Discovery
- WES based Biomarker Discovery
- Personal Tumor-Specific Neoantigen Vaccine Development
- MRD Monitoring
- Microsatellite Instability Analysis
- One-Stop Cancer 3D Modeling
- Circle-Seq based eccDNA Identification
- RNA-Seq based Tumor Microenvironment Analysis