Knowledge-based methods are of value in the prediction of receptor structures to aid the design of drugs, herbicides or pesticides, antigens in vaccine design, and novel molecules in protein engineering. Creative Biolabs is dedicated to establishing the most exquisite service platform for our clients and our one-stop protein engineering services can provide comprehensive technical support for advancing our clients' projects.
The design of the tertiary structures of proteins may be carried out using a knowledge-based approach. This depends on the identification of analogies in secondary structures, motifs, domains or ligand interactions between a protein to be modeled and those of known three-dimensional structures. The approach simultaneously aligns the known tertiary structures, selects fragments from the structurally conserved regions on the basis of sequence homology, aligns these with the ‘average structure' or ‘framework', builds on the loops selected from homologous proteins or a wider database, substitutes sidechains and minimizes the energy of resultant model.
Knowledge-based methods have been used to design a variety of constructs: transmembrane binding peptides, surface tissue peptide superstructures and protein crystals. For example, the computed helical anti-membrane protein (CHAMP) protocol was capable of designing α- and β-helical peptides that specifically bind to the transmembrane (TM) helix of a target protein. Scientists used known membrane-protein structures to create backbone templates and a membrane depth-dependent knowledge-based potential to sample the “relevant” sequence and rotamer space. This strategy has resulted in several designs whose combination has been experimentally verified. The specificity and efficacy of CHAMP peptides interacting with their target integrins were demonstrated via a biological activity assay. Designed CHAMP peptides specifically interact with, and thus stimulate the activity of only the integrin whose transmembrane domain they were designed to interact with.
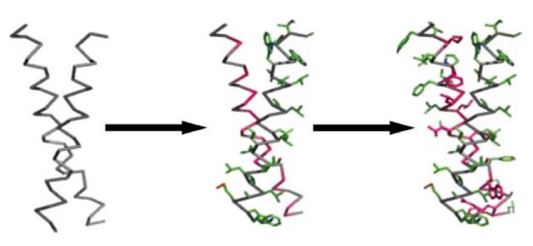 Fig.1 The CHAMP design process.
Fig.1 The CHAMP design process.
Protein design approaches have revealed many fundamental features of protein structure and stability. Knowledge-based protein design is fundamental to structural biology and drug discovery. There is an urgent request for computational methods capable of reliably generating and analyzing protein structure. We can provide knowledge-based protein design to meet customers' specific requirements.
Creative Biolabs has been involved in the field of protein design for many years and we are committed to completing your project with high quality. We have accumulated a wealth of scientific experience from our completed projects and provide you with the best services to ensure your requirements are met. If you are interested in our services, please contact us for more details.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.