Complementary DNA (cDNA) libraries are an important tool in biological research. cDNA has no introns and can be expressed in prokaryotic cells. Therefore, cDNA libraries are commonly used to express eukaryotic genes in prokaryotes. cDNA libraries are most useful in reverse genetics. In addition, cDNA libraries are often used for functional cloning to identify genes based on the function of the encoded protein. When studying eukaryotic DNA, expression libraries are constructed using complementary DNA (cDNA) to help ensure that the inserted fragment is truly a gene. Given the various functions of cDNA libraries, Creative Biolabs offers a comprehensive cDNA library construction service to meet your research requirements.
The investigators used Nb.BbvC to construct the cDNA library. Nb.BbvCI recognizes sequences seven bases long and therefore rarely digests cDNAs. Even if a cDNA contains more than two recognition sites, in cases where the distance between the two sites is relatively small (within about 30 bp), the nicked fragment will dissociate during denaturation. Therefore, the digestion of cDNAs with this enzyme is extremely rare. Before constructing the library, the investigators first performed experimental experiments in which the GUS coding sequence (GUS) was inserted into pER8 using nickase.
The study strategy is shown in Fig 1. The vector and the GUS coding sequence were amplified by PCR using the indicated primers and Q5 DNA polymerase. 50 µl of the reaction mixture containing 100 ng of pER8 or pBI221 was first incubated at 96°C for 1 min, followed by 10 cycles of denaturation at 98°C for 20 s, annealing at 60°C for 30 s, elongation at 72°C for 12 min, and finally extension at 72°C for 10 min. The different sequences (NL and NL2) of the ends of vectors and GUS were used for annealing. The primer sequences contained the recognition sequence of Nb.BbvCI (GCT GAG G). DpnI was also added to the reaction to digest the template DNA, since the template DNA recovered from E. coli cells is methylated and DpnI specifically digests methylated DNA. The short single-stranded fragments produced by Nb.BbvCI digestion can be easily dissociated by incubation at high temperature (72°C) to produce fragments with single-stranded ends. The vector and GUS fragments are mixed in a 1:2.5 molar ratio and annealed overnight at 60°C in a buffer containing 10% polyethylene glycol (PEG), 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM EDTA to enhance annealing.
The resulting DNA solution was transformed into ECOS-capable DH5α. The pER8::GUS generated after annealing was also transformed into the competent cells. Based on the results of the study, the cloning efficiency was calculated to be 11%. In addition, all of the 16 randomly selected colonies carried plasmids of GUS, indicating that the cloning rate without the insert was less than 6%.
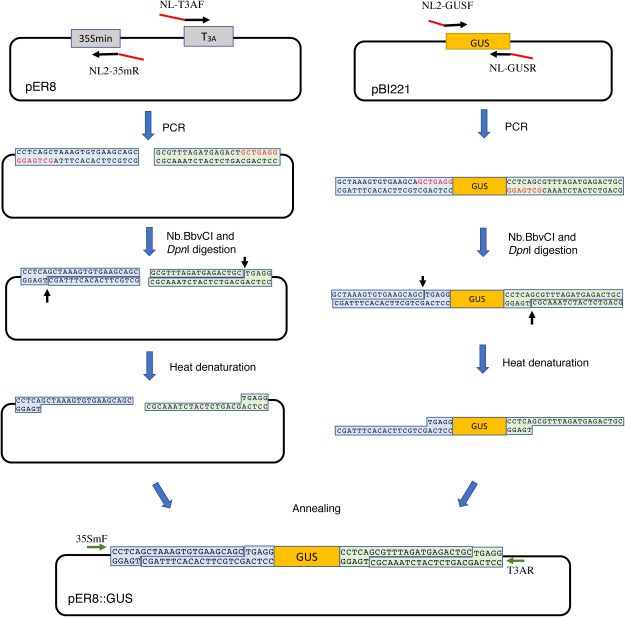 Fig 1. Schematic diagram and flowchart of NE-LIC. (Nishioka, D., et al., 2020)
Fig 1. Schematic diagram and flowchart of NE-LIC. (Nishioka, D., et al., 2020)
Using this strategy, a cDNA library was constructed in an estrogen-inducible vector, pER8. Linearized pER8 with linkers at both ends was amplified using primers NL-T3AF and NL2-35MR, and then digested with DpnI to minimize the amount of circular DNA, which could result in background clones without inserts. The primers NL2-pSKF and NL-pSKR were used to amplify the cDNA library constructed in pSK34 previously. PCR conditions were identical to those in the experiments using GUS sequences. Next, 2 µg of vector DNA and cDNA were mixed, treated with Nb.BbvCI for 2 h at 37°C, and then incubated for 15 min at 72°C. After fractionation with Sephacryl S-400 spin columns to remove the short fragments and purification with a silica membrane-based DNA purification kit, the mixture of vector DNA and cDNA was incubated overnight at 60°C in the above buffer. The annealed DNA was transformed into E. coli electrophoresis cells to produce 5.1×105 independent colonies. Fig 2. shows the results of colony PCR from the transformed colonies. All 16 colonies carried plasmids with cDNA inserts in the length range of 500 bp to 2,000 bp, and no clones without inserts were detected. Thus, the cDNA library was successfully constructed in pER8. The results suggest that nicking enzyme-based plasmid constructs can be a powerful alternative to exonuclease-based constructs because of their low insertion-free background. This approach can be used to construct cDNA libraries by synthesizing cDNAs from mRNA using the SMART technique, although the investigators amplify cDNAs from a preexisting cDNA library by PCR and insert them into another vector using NE-LIC.
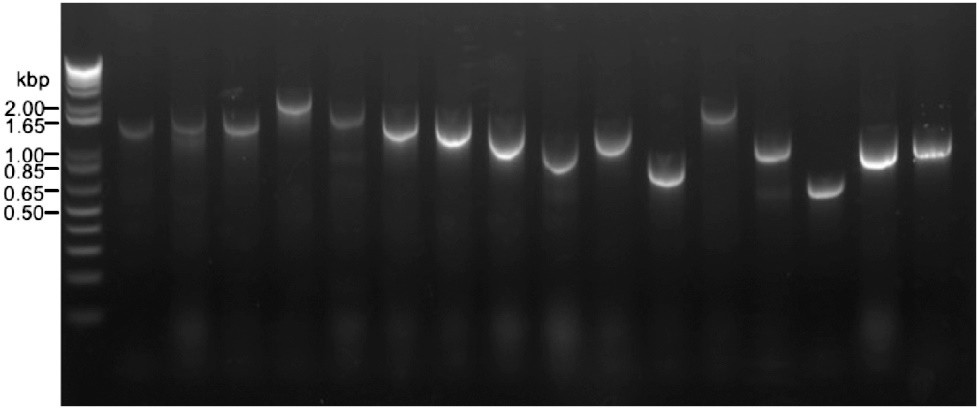 Fig 2. Inserts detected by colony PCR in the cDNA library in pER8. (Nishioka, D., et al., 2020)
Fig 2. Inserts detected by colony PCR in the cDNA library in pER8. (Nishioka, D., et al., 2020)
Reference
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.