- Home
- Resources
- Knowledge Center
- Protocols
- Conjugation Site Analysis of Lysine-Conjugated ADCs
Conjugation Site Analysis of Lysine-Conjugated ADCs
Lysine-conjugated antibody-drug conjugates (ADCs) are formed by attaching cytotoxic drugs to reactive lysine residues of monoclonal antibodies (mAbs) through chemical linkers. During production, the payloads are conjugated nonspecifically to lysine residues in mAbs, resulting in a heterogeneous mixture of ADCs with both different numbers and conjugation sites of drug payloads per mAb. On account of the drug conjugation sites and levels that both have significant influences on the physical and pharmaceutical properties of ADCs, a reliable and straightforward approach for conjugation site analysis for ADCs is in high demand.
This article takes a lysine-conjugated ADC, Trastuzumab-MCC-DM1 (T-DM1), as a model ADC to briefly describe a strategy for how to identify and quantify conjugation sites, aiming to provide researchers with universally applicable methods to lysine-conjugated ADCs for the recognition of conjugation sites and the measurement of conjugation levels.
Disclaimer
This procedure is only a guideline. Please note that Creative Biolabs is unable to guarantee experimental results if it is operated by the customer.
Preparation of ADC Digested Peptides
Materials:
Denaturation buffer: 8 M urea in 50 mM NH4CO3 buffer.
Reduction solution: 0.5 M dithiothreitol (DTT) in 50 mM NH4CO3 buffer.
Alkylation solution: 0.4 M iodoacetamide (IAA) in 50 mM NH4CO3 buffer.
Chymotrypsin at 20 μg/mL was dissolved in 50 mM acetic acid.
10% formic acid (FA).
Activation solution for desalting: ACN with 0.1% FA.
Equilibrium solution for desalting: ultrapure water with 0.1% FA.
Eluent solution for desalting: ACN-water (1:1, v/v) with 0.1% FA.
Vacuum concentrator.
Original T-DM1 and generic T-DM1 freeze-dried powder injection.
Procedure:
1. Dissolve the T-DM1 freeze-dried powder injection in water to prepare the ADC sample solution.
2. Transfer 10 μL of ADC sample solution to 100 μL of 8 M urea and briefly vortex the mixture. Add 2 μL of 0.5 M DTT and incubate the solution in 56 °C water bath for 30 min.
3. Add 10 μL of 0.4 M IAA and incubate in the dark at 37°C for another 30 min, then add 2 μL of 0.5 M DTT and incubate at RT for 15 min (see Note 1).
3. Add chymotrypsin to the T-DM1 sample at a ratio of 1:50 for overnight digestion at room temperature.
4. Add 10% FA to acidify the digested peptide sample to 0.1% FA.
5. The above digestive peptide is eluted by the dehydrated column, and the eluted fraction is collected in a tube.
6.
Dry the eluted peptides using a pressure-blowing concentrator with N2. Store the peptide samples at -80 °C until use.
Note:
1. DTT and IAA should be freshly prepared and IAA must be kept away from light.
Identification of Conjugation Sites
Materials:
Mobile phase A: LC-MS grade water with 0.1% FA.
Mobile phase B: LC-MS grade ACN with 0.1% FA.
A nano-scale liquid chromatography (nanoLC) system
A reversed-phase C18 nano-column (75 μm ×150 mm, 3 μm particle size) for peptide separation.
Procedure:
1. Redissolve the ADC digestion samples with 20 μL 0.1% FA in LC-MS grade water, and centrifuge the samples at 18,000 ×g for 10 min at 4 °C.
3. Transfer 15 μL of supernatant to sampling vials for nanoLC-MS/MS analysis.
3. Set up the following elution gradient:
5% B for 4 min, 5–60% B for 56 min, 60–95% B for 15 min, 95% B for 10 min, 95–5% B for 5 min, and 5% B for 10 min equilibration.
4. Set up the flow rate at 300 nL/min.
5. nanoESI-MS/MS Detection.
An electrospray ionization (ESI) source operated in positive mode is used to introduce positively charged peptide ions.
The information-dependent acquisition (IDA) mode was employed for peptide mapping and subsequent identification of drug-conjugated lysine. The scan parameters under IDA mode were set as follows: the m/z range of 350–1500 Da for TOF-MS scan, the m/z range of 100–2500 for product ion scan, and declustering potential (DP) as 100 V.
Conjugated Peptide Screening
Procedure:
1. The software ProteinPilot was used for peptide mapping and DM1-conjugated site identification.
2. Input the heavy chain (HC) and light chain (LC) sequences of the antibody, trastuzumab, to a fasta file, then input the PTMs that are the DM1 conjugation and also the mass shift caused by drug-linker conjugation as modifiers. All the precursors and specific fragment ions of lysine-containing peptides carrying an appropriate mass shift are assigned as DM1-conjugated peptides (see Note 2).
Note:
1. The conjugation of cytotoxic drugs to antibodies leads to incremental masses of precursors and fragment ions corresponding to DM1 conjugation for peptides that carry such modification. Drug conjugation leads to the creation of SFIs, which are highly abundant in MS/MS spectra. Consequently, these SFIs were utilized in this case as a reliable benchmark to eliminate false positive hits from the results of database searches.
Quantitative Analysis of Lysine Conjugation Levels
Materials:
LC-MS/MS mobile phase A: ultrapure water with 0.1% FA.
LC-MS/MS mobile phase A: ACN with 0.1% FA.
HPLC system
C18 column (2.1 × 150 mm, 5 μm, 300 A) for peptide separation.
DM1 and lys-MCC-DM1 were dissolved in MeOH at 1 mg/mL.
High-speed freezing centrifuge.
Procedure:
1. Digest TDM1 and desalt the digested peptides as described in the above protocol of preparation for the ADC digested peptides.
2. Reconstitute the digested sample in aqueous 0.1% FA buffer to a concentration of approximately 1 mg/mL.
3. Aliquot the sample and dilute the aliquots to 2.5-, 5-, 10-, and 25-fold, and set up five concentrations to calculate the relative ionization intensity factor (RIIF) for identified DM1-conjugated and unconjugated peptides.
4. Add a peptide standard to each sample as an internal standard (IS).
5. Centrifuge serially diluted samples at 30,000 × g for 5 min at 4 °C and transfer the supernatant of 5 μL to sampling vials for LC-MS analysis.
6. Set up the following analysis gradient:
5% B for 1 min, 5–65% B for 42 min, 65–90% B for 2 min, 90% B for 7 min, 90–5% B for 2 min, 5% B for 6 min.
7. The flow rate is set at 0.2 mL/min.
8. Run and record the pseudo-multiple reaction monitoring (MRM) responses for each monitored peptide and quantify the corresponding peak area values collected from samples of serial concentrations (see Note 3).
9. Make the concentration-response curves for all the conjugated and unconjugated peptides, and then calculate the corresponding slope rates.
10. Calculate the normalized conjugation ratios for all the identified DM1 conjugation peptides to obtain the conjugation level of each modified lysine residue by using the following equations:
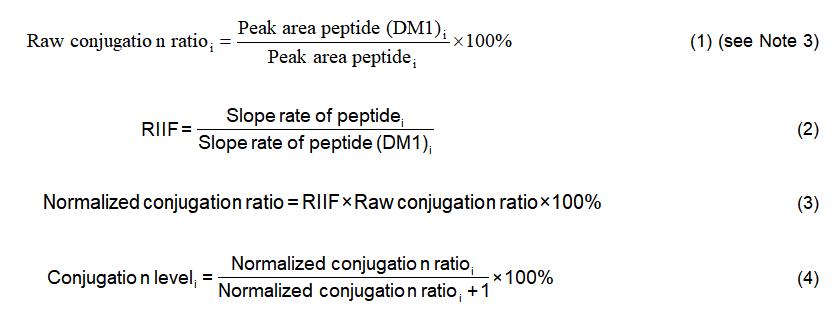
11. Add up the conjugation level for each site. The summed value should be close to the drug-antibody ratio (DAR) acquired by measuring the intact masses of the conjugated and naked mAbs (see Note 4).
Note:
1. Peptides have multiple charges and, thus, multiple m/z values. The charges of conjugated and unconjugated peptides chosen to monitor for MRM analysis are identical.
2. Make sure the m/z (among different charge states) values of unconjugated and conjugated peptide pairs one chooses for raw conjugation ratio calculation are the same as those one chooses for normalized conjugation ratio calculation.
For Research Use Only. NOT FOR CLINICAL USE.

Online Inquiry
Welcome! For price inquiries, please feel free to contact us through the form on the left side. We will get back to you as soon as possible.
