NAA Paratope Mapping
Paratope mapping technology represents an attractive method to investigate the binding sites of antibody recognized by antigen. It has been used in the prediction of vaccine, the detection of the immune response and autoimmunity, etc. With perfect strategies and abundant experience, Creative Biolabs can provide a comprehensive range of natural autoantibodies (NAA) services. Our expert team can offer the most professional solutions and the advanced technologies which will ensure the progress of our customers’ research. Aided by extensive experience in antibody analysis and detection, our high-quality NAA services will promote the treatment research of various diseases.
Overview
The Importance of NAA Paratope Mapping
Paratope mapping is a method to characterize the binding sites, or paratopes of antibodies recognized by antigens. During the development of vaccines and biotherapeutics, one of the most critical processes is the identification and analysis of the antibody paratopes. There are a variety of important applications of paratope mapping in the antibody, in brief, including the prediction of suitable antigens as vaccine components, the exploration of the immune response and autoimmunity, the characterization of the therapeutic antibody’s mechanism of action.
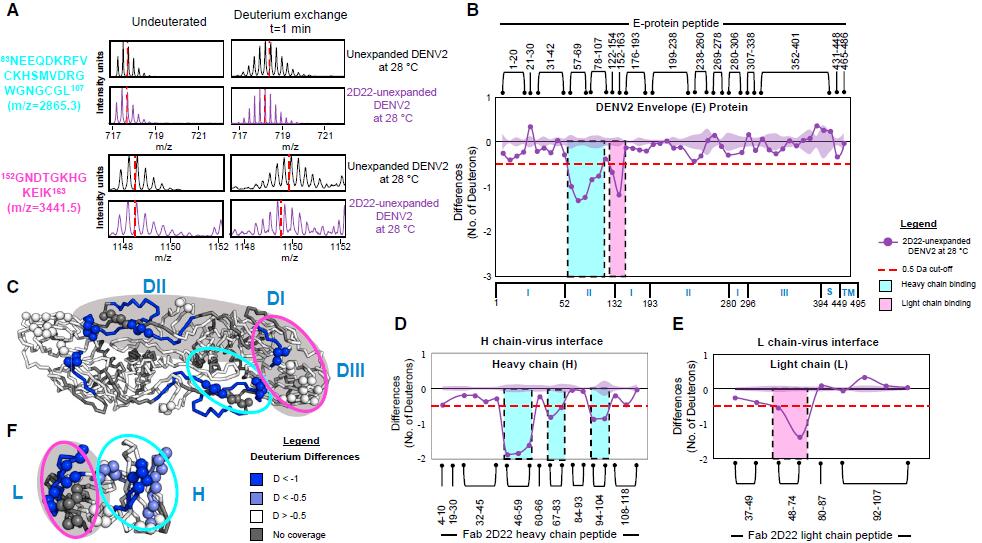 Fig.1 Epitope and paratope mapping of 2D22 unexpanded DENV2 complex (2D22-DENV2UN) at 28C by amide hydrogen/deuterium exchange mass spectrometry (HDXMS). (Lim, 2017)
Fig.1 Epitope and paratope mapping of 2D22 unexpanded DENV2 complex (2D22-DENV2UN) at 28C by amide hydrogen/deuterium exchange mass spectrometry (HDXMS). (Lim, 2017)
Service Category
Methods of Paratope Mapping
The methods of NAA paratope mapping provided by Creative Biolabs include but not limited to the followings:
-
Computational Procedure
The computational procedure is composed of rigid body docking, free and steered molecular dynamics (MD) simulations and homology modeling. These key paratopes residing on the target antibody and their partners on antigen can be identified using the computational procedure. Macromolecular docking is the computational model of the quaternary structure of complexes consisted of more than two biological macromolecules which interact with each other. Molecular dynamics (MD) means the analysis of the motion of molecules and atoms. Molecules and atoms are performed to interact for a certain time for the overview of the dynamical evolution of this system. Homology modeling builds an atomic-resolution model of the protein from its amino acid sequence, and furthermore builds an experimental three-dimensional structure of a correlative isogenous protein. -
Hydrogen-deuterium Exchange and Mass Spectrometry
In hydrogen-deuterium exchange and mass spectrometry, the hydrogen atoms in the solvent will be exchanged by backbone amide hydrogen atoms and if there is deuterium in the solvent but don’t contain hydrogen, the same exchange process occurs, thus, the protein can be deuterated. The flexibility and availability of the target part of the protein decide the rate of any special hydrogen atom happens. The hydrophobic core of a rigid protein where the hidden hydrogen atoms will be exchanged at all, while those hydrogen atoms on a flexible loop will be exchanged nearly instantaneously. -
Peptide-based Approaches
Peptide-based approaches are based on the overlap peptides of the sequence of the antigen. Following the peptides fixated onto a solid surface as an array, the binding to the antibody is determined in an ELISA format. This method can facilitate screening of antibodies with very huge phage libraries available. In addition, the recombinant peptide genes from binding phage particles can be sequenced to identify the sequences and putative epitopes.
Key Features
Features of Our Services
- High resolution
- Less sample amount required
- Used for both linear and conformational epitopes
- High-efficient and cost-effective
Resource
Creative Biolabs is a professional NAA services provider to help worldwide clients to detect NAA repertoire, perform NAA profiling, and identify NAA epitopes or paratopes, depending on a series of novel methods and tools. Our high-quality services will contribute greatly to the success of your projects. Please feel free to contact us for more information and a detailed quote.
Reference
- Lim, X.X.; et al. Epitope and paratope mapping reveals temperature-dependent alterations in the dengue-antibody interface. Structure. 2017.

