MRM/PRM Targeted Exosomal Proteomic Quantitative Service
Overview Services Features FAQs
Creative Biolabs offers a high-throughput and highly accurate MRM/PRM targeted exosomal proteomic quantitative service that can validate these exosome-based markers in large biological sample sizes based on differential proteomic data.
Introduction of MRM/PRM Targeted Exosomal Proteomic Quantitative
-
Targeted quantitative proteomics uses mass spectrometers for ion screening and targeted detection of fragmented ions from selected exosomal peptides.
-
There are now two primary technical paths that are being followed: PRM (Parallel Reaction Monitoring) and MRM (Multiple Reaction Monitoring).
-
Using triple-quadrupole mass spectrometers, MRM is intended for the focused examination of certain proteins, peptides, or tiny compounds in complicated materials.
-
PRM uses high-resolution/precision mass spectrometers and is a simplified version of MRM.
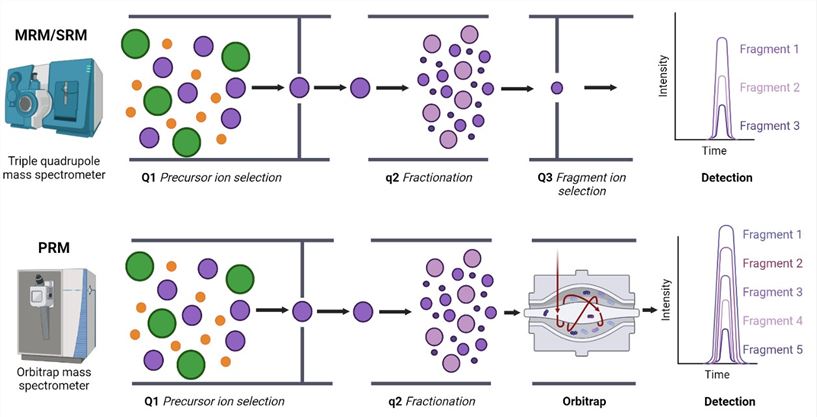 Fig. 1 MRM/SRM and PRM targeted proteomics.1
Fig. 1 MRM/SRM and PRM targeted proteomics.1
MRM/PRM Targeted Exosomal Proteomic Quantitative Service at Creative Biolabs
We offer one-stop MRM/PRM exosome quantitative protein analysis services, including MS analysis, raw data analysis, bioinformatics analysis, specific peptide selection, isotope-labeled peptides, and MRM/PRM method establishment and optimization, based on the target protein information you provide for the desired study. There are several facets of bioinformatics analysis available, including
-
Spectrogram analysis, and protein data quality assessment.
-
PCA analysis of multi-component samples.
-
Functional annotation of identified proteins: GO functional annotation, KEGG functional annotation, and COG functional annotation.
-
Venn diagram and volcano plot statistical analysis of proteins with differential expression.
-
K-means clustering and hierarchical clustering for the study of proteins with differential expression.
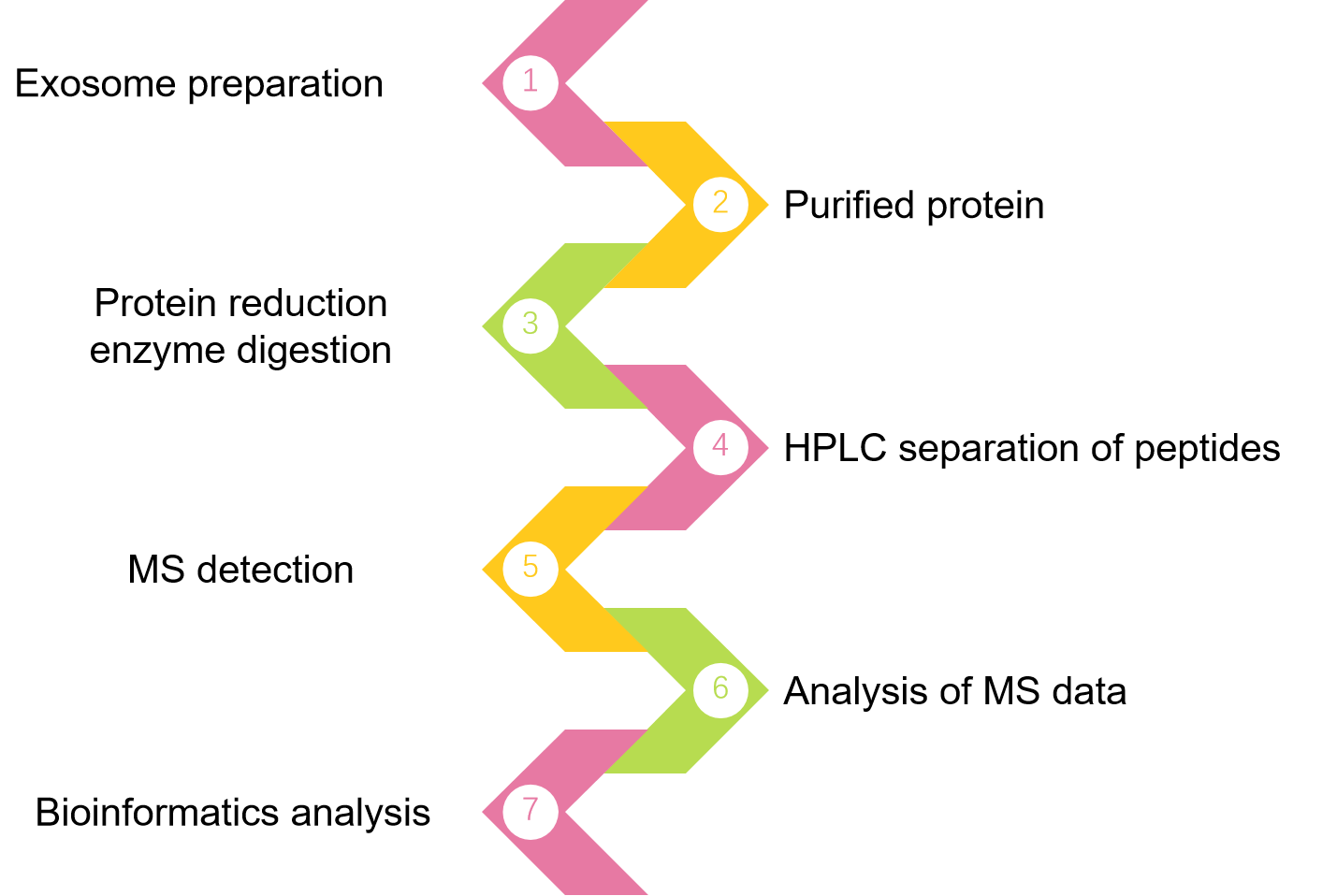 Fig. 2 MRM/PRM workflow.
Fig. 2 MRM/PRM workflow.
Features and Applications of MRM/PRM
1. MRM assay
-
Utilizes a triple quadrupole mass spectrometer for targeted large-molecule protein detection.
-
Benefits:
Reduces interference from other ions.
Improves signal-to-noise ratio, sensitivity, and reproducibility.
Suitable for high-throughput monitoring of marker proteins.
-
Can achieve absolute quantification by adding isotopically labeled peptides as internal references.
-
Optimal for measuring particular sets of target exosomal proteins in large-scale specimens.
2. PRM assay
-
Relies on high-resolution mass spectrometers for targeted large-molecule protein detection.
-
Features:
Single mass spectrometer for proteome screening and validation.
Easy operation and low detection cost.
Can target a large number of ions for monitoring.
-
Challenges:
Instrumentation can be less stable due to nanoliter liquid phase system.
Requires fine adjustment of MS acquisition parameters for accuracy and precision.
-
Typically used shortly after non-targeted proteomics and before mass spectrometer maintenance.
-
Appropriate for exosomal protein samples in tiny batches that necessitate protein-targeted quantitative assays.
FAQs
Q: What is MRM/PRM Service?
A: MRM/PRM Targeted Exosomal Proteomic Quantitative Service is a specialized service that utilizes MRM or PRM techniques for targeted analysis of exosomal proteins. This service provides accurate and quantitative measurement of specific proteins in exosomes, enabling detailed insights into exosome biology and potential biomarker discovery.
Q: What are the advantages of using MRM/PRM for exosomal proteomic analysis?
A: Targeted protein quantification can be achieved with great precision, repeatability, and sensitivity when using MRM/PRM methods. These techniques allow for the simultaneous measurement of multiple proteins in a single analysis, making them ideal for comprehensive and quantitative analysis of exosomal proteomes.
Q: How do I place an order for the service?
A: To place an order or to inquire about our services, please contact our customer support team and fill out the online inquiry form on our website. We will be happy to assist you with your inquiry and provide further information about our services.
Using high-resolution, high-precision mass spectrometry, Creative Biolabs provides MRM/PRM focused exosomal proteomic quantitative services that facilitate the selective identification and accurate quantification of exosomal proteins. Kindly get in touch with us for a quote.
Reference
-
Guerrero, Laura, Alberto Paradela, and Fernando J. Corrales. "Targeted Proteomics for Monitoring One-Carbon Metabolism in Liver Diseases." Metabolites 12.9 (2022): 779. Under Open Access license CC BY 4.0. The image was modified by revising the title.
For Research Use Only. Cannot be used by patients.
Related Services:

 Fig. 1 MRM/SRM and PRM targeted proteomics.1
Fig. 1 MRM/SRM and PRM targeted proteomics.1
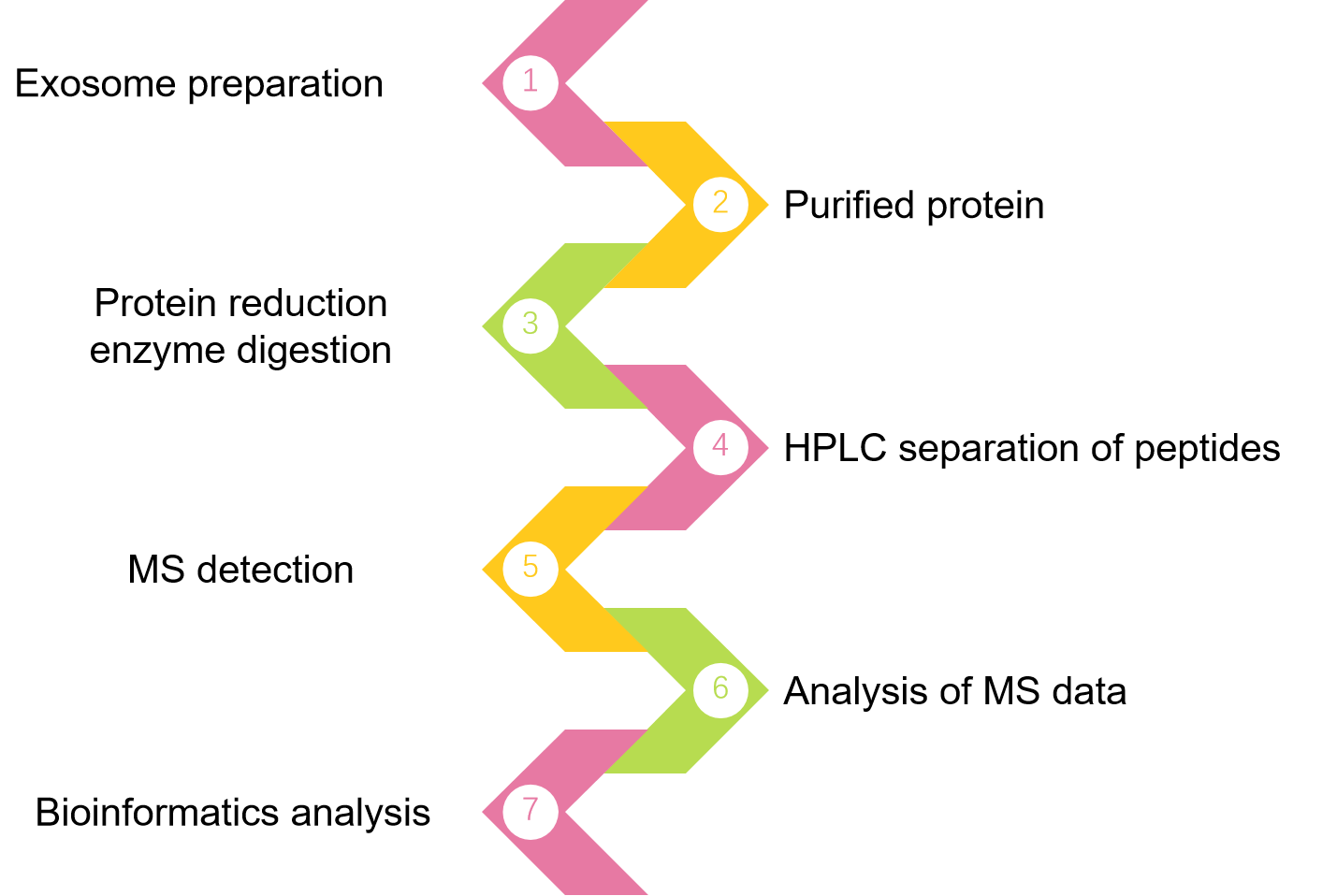 Fig. 2 MRM/PRM workflow.
Fig. 2 MRM/PRM workflow.