Phosphorylated Exosomal Proteomic Detection Service
Overview Services Features FAQs
Overview
Exosomes are membrane-coated nanoparticles that protect their internal contents from external proteases and other enzymes, offering a new solution for analyzing phosphorylated proteins. Leveraging advanced proteomics analysis platforms, Creative Biolabs introduces its exosome phosphorylation proteomics analysis service.
Background of Protein phosphorylation
Protein phosphorylation is a critical and widespread molecular regulatory mechanism that influences nearly all aspects of cell function. The state of phosphorylation events can provide important clues about disease conditions. However, few phosphoproteins have been developed as disease markers due to the invasive nature of tissue biopsies and the highly dynamic nature of protein phosphorylation in typical long and complex biopsies. This makes the detection of phosphoproteins in tissues particularly challenging.
Significance of Proteomic Analysis of Exosomal Phosphorylation
The discovery of exosomes has provided solutions to these challenges. Exosomes' vesicular properties make them highly stable in biological fluids over extended periods, creating the potential to develop phosphorylated proteins for medical diagnostics. Detecting genome output (active proteins, especially phosphorylated proteins) offers more direct and real-time information about biological functions and disease progression, especially in cancer.
Services
Phosphorylation Proteomics Service for Exosomes
We can currently isolate and identify phosphoproteins from exosomes and use immobilized metal ion affinity chromatography (IMAC) to efficiently increase the number of S, T, and Y phosphorylated peptides. Additionally, using Data-Independent Acquisition (DIA) technology, we can collect all signals within the mass spectrum scanning range without bias, effectively addressing the challenges of qualitative and quantitative phosphorylation modification.
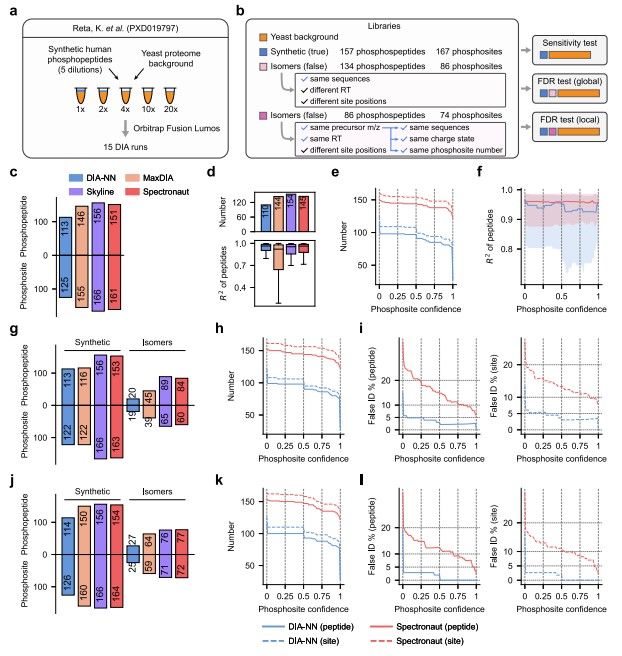 Fig.1 Evaluation of phosphopeptide identification and phosphosite localization.1,2
Fig.1 Evaluation of phosphopeptide identification and phosphosite localization.1,2
Features
-
Full Scan Mode: The DIA mode significantly reduces interference from high abundance signals, comprehensively collecting high and low abundance signals and increasing data coverage by nearly 40%.
-
Ultra-High Reproducibility and Stability: The repetition rate for large sample identification has increased by nearly 40%, and the quantitative precision has nearly doubled.
-
Ultra-High Quantitative Accuracy: Our quantitative ability is close to the gold standard of multiple reaction monitoring (MRM), also known as Selective Reaction Monitoring (SRM) targeting technology.
Creative Biolabs is dedicated to the study of exosomes and provides comprehensive services for exosome research. Our phosphorylated proteomics analysis service brings the development of exosomal phosphoproteins as potential disease markers within reach. If you are interested in our services, please contact us for more information.
FAQs
Q: How much exosome sample do I need to provide for phosphoproteomics analysis?
A: For phosphoproteomics analysis, we require a minimum of 1mL of exosome sample, with a concentration of ≥10^10 particles/mL if quantified by particle number, or ≥0.25μg/μL if quantified by BCA. Providing sufficient sample volume ensures accurate and reliable results.
Q: What is the turnaround time for phosphoproteomics analysis of exosome samples?
A: The turnaround time for phosphoproteomics analysis of exosome samples typically ranges from 8 to 10 weeks. This timeframe includes sample preparation, data acquisition, and detailed analysis. We aim to provide comprehensive and accurate results within this period.
Q: What kind of information will I receive from the phosphoproteomics analysis of my exosome samples?
A: From the phosphoproteomics analysis, you will receive comprehensive information on the phosphorylation status of proteins within your exosome samples. This includes the identification of phosphorylated proteins, the specific phosphorylation sites, and quantitative data on phosphorylation levels. The results will provide valuable insights into the signaling pathways and biological processes regulated by exosomal phosphoproteins.
References
-
Lou, R.; et al. Benchmarking commonly used software suites and analysis workflows for DIA proteomics and phosphoproteomics. Nature Communications. 2023, 14(1):94.
-
Distributed under Open Access license CC BY 4.0, without modification.
For Research Use Only. Cannot be used by patients.
Related Services:

 Fig.1 Evaluation of phosphopeptide identification and phosphosite localization.1,2
Fig.1 Evaluation of phosphopeptide identification and phosphosite localization.1,2