Streptococcus pneumoniae-derived Exosome Research & Application
Overview Workflow Exosome Insights Research Support Key Benefits Testimonials FAQs
Introduction & Scientific Context
Exosomes secreted by Gram-positive bacteria are gaining attention as an emerging field of microbiology and host–pathogen interaction research. Among them, vesicles from Streptococcus pneumoniae (S. pneumoniae) are of particular interest because of the organism 's role as a widely studied respiratory pathogen and commensal bacterium. Research on pneumococcal vesicles has revealed that they are not random cellular by-products but rather structured carriers of proteins, nucleic acids, and lipids. These vesicles are now recognized as important mediators of microbial communication, immune modulation, and interspecies signaling.
For researchers interested in host–pathogen relationships, immune activation pathways, and vesicle-based vaccine design, S. pneumoniae-derived exosomes represent a promising yet complex system. Creative Biolabs supports laboratories worldwide in exploring these vesicles by offering tailored workflows for vesicle generation and optional advanced analyses depending on available strain libraries. By collaborating with Creative Biolabs, investigators gain access to reliable processes and technical expertise designed to ensure that data from pneumococcal vesicle studies are reproducible and credible.
Customized Workflow for Vesicle Preparation
The preparation of pneumococcal vesicles requires careful handling, given the biological complexity of the species and its fastidious growth characteristics. At Creative Biolabs, the standard exosome development workflow has been optimized for consistency. Importantly, while the basic pipeline focuses on isolation and vesicle production, optional downstream analyses such as proteomic characterization, RNA sequencing, or functional immunoassays can be requested (availability dependent on strain-specific resources).
Standardized steps include:
-
Culturing S. pneumoniae overnight at 37 °C under controlled aeration.
-
Centrifugation to remove bacterial cells and residual biomass.
-
Sterile filtration of the culture supernatant, followed by sterility validation.
-
Ultracentrifugation to pellet pneumococcal exosome-like vesicles.
-
Resuspension in PBS and preservation for experimental application.
-
(Optional) Size-exclusion chromatography for further purification, depending on downstream research needs.
This flexible system allows Creative Biolabs to deliver vesicles as raw isolates for labs seeking to perform their own assays, or as highly purified material (optional) where detailed vesicle content analysis is required.
Biological Insights from S. pneumoniae-derived Exosomes
The following table summarizes selected findings from global research efforts on pneumococcal vesicles. These studies do not reflect results generated by Creative Biolabs but instead compile the broader scientific progress in the field, showing how exosomes from S. pneumoniae contribute to bacterial survival and host interactions.
|
Research Focus
|
Findings and Implications
|
|
Structural Heterogeneity
|
Cryo-EM analyses show that S. pneumoniae vesicles are morphologically diverse, ranging from spherical to chain-like forms, suggesting functional diversity in their biological roles.
|
|
Cell Compatibility
|
In vitro viability tests indicate that human epithelial cells, keratinocytes, macrophages, and dendritic cells tolerate vesicles without cytotoxic effects, underscoring their potential use as model delivery vehicles.
|
|
Uptake by Host Cells
|
Fluorescent labeling studies demonstrate that immune cells, particularly macrophages, efficiently internalize pneumococcal vesicles, pointing to targeted delivery of bacterial components.
|
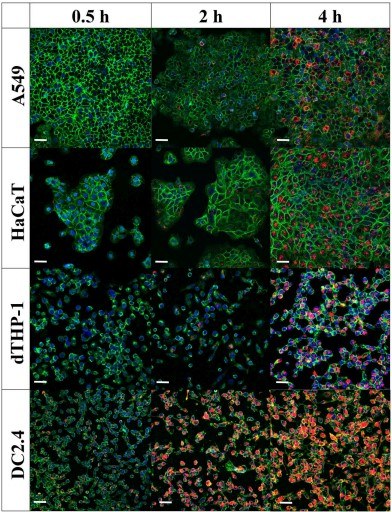 Fig.1 Cellular uptake of Streptococcus pneumoniae-derived exosomes increases with time.1
Fig.1 Cellular uptake of Streptococcus pneumoniae-derived exosomes increases with time.1
|
Research Focus
|
Findings and Implications
|
|
Stimulation of Immune Pathways
|
Vesicles activate NF-κB signaling and promote release of pro-inflammatory cytokines in dendritic cells and macrophages, highlighting their immunostimulatory properties.
|
|
Systemic Immune Effects
|
Mouse studies show that vesicles increase M2 macrophage populations and modulate adaptive immunity, suggesting a role in shaping chronic bacterial carriage.
|
|
Adaptive Immune Recruitment
|
Inoculation models reveal recruitment of host immune cells to local tissues after vesicle exposure, providing insights into host colonization dynamics.
|
|
Role of Lipoproteins
|
Mutant strain analyses confirm that pneumococcal lipoproteins embedded in vesicles are key triggers for NF-κB activation and immune signaling regulation.
|
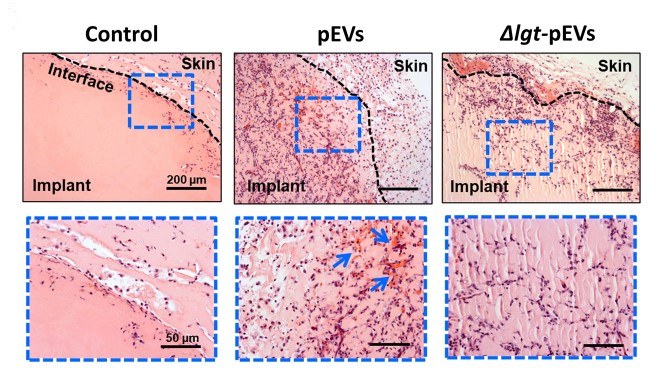 Fig.2 Streptococcus pneumoniae-derived exosomes facilitated host cell recruitment.2
Fig.2 Streptococcus pneumoniae-derived exosomes facilitated host cell recruitment.2
This growing body of literature positions pneumococcal exosomes not only as byproducts of bacterial physiology but also as active agents influencing immune responses and infection pathways.
Creative Biolabs ' Dedicated Research Support Platform
Creative Biolabs has established a specialized infrastructure for Gram-positive bacterial vesicle research. The platform integrates customizable vesicle preparation, scalable production capacity, and optional molecular profiling tools. By leveraging this platform, scientists can conduct controlled experiments on pneumococcal vesicles without the variability associated with in-house isolation attempts.
Flexibility
Scalability
Consistency
Expertise
Flexibility
Basic vesicle isolation services with optional proteomics, lipidomics, or RNA analysis (library-dependent).
Scalability
Ability to deliver vesicles in research-ready volumes suitable for pilot studies through to multi-assay projects.
Consistency
Standardized SOPs minimize experimental variability, enabling comparable results across independent studies.
Expertise
Technical consultation on vesicle preparation strategy and design of exploratory workflows.
Distinctive Benefits of Working with Creative Biolabs
Collaborating with Creative Biolabs offers several advantages for academic and industrial research groups aiming to deepen their investigation into pneumococcal vesicles:

Reliability
All preparations follow rigorously tested protocols to ensure reproducibility.

Optional Enrichment
Advanced characterization assays can be integrated into projects depending on the availability of strain libraries.

Knowledge Base
Years of accumulated expertise in Gram-positive exosome systems help clients interpret results with context.

Customizability
Project scopes are designed to match each laboratory 's requirements, whether for preliminary exploration or detailed molecular study.
By combining technical expertise with flexible service design, Creative Biolabs stands as a trusted partner for teams seeking meaningful outcomes in bacterial vesicle research.
Feedback from Research Collaborators
Scientific collaborators consistently highlight Creative Biolabs ' professionalism and reliability in delivering vesicle-based research support:
"Creative Biolabs provided precisely what our project required—a consistent supply of well-prepared pneumococcal vesicles. Their attention to sterility and reproducibility allowed us to move forward with confidence."
"What stood out most was Creative Biolabs ' adaptability. We started with standard vesicle isolation but later expanded into proteomic profiling once preliminary data looked promising. Having that flexibility was invaluable."
"The technical consultation provided by Creative Biolabs saved us significant time. Their team 's prior knowledge of Gram-positive vesicles helped us design experiments more effectively than working in isolation."
As the field of bacterial vesicle research continues to expand, S. pneumoniae exosomes remain an important focus for understanding host–pathogen dynamics and for developing new research tools. Creative Biolabs remains committed to advancing this area by providing rigorous workflows, optional downstream services, and collaborative expertise. For research teams eager to investigate the structural, functional, and immunological features of pneumococcal vesicles, Creative Biolabs offers a dependable platform to accelerate progress. Researchers are encouraged to reach out to discuss specific requirements, whether the goal is basic characterization or hypothesis-driven exploration of exosome-mediated interactions.
FAQs
Q: How do Streptococcus pneumoniae exosomes contribute to bacterial pathogenesis?
A: These exosomes are implicated in enhancing bacterial virulence by facilitating the delivery of pathogenic factors that can manipulate host defenses. For example, they can aid in the evasion of the host immune response and alter the local microenvironment to favor bacterial survival and proliferation.
Q: What research methods are used to study Streptococcus pneumoniae exosomes?
A: Researchers typically employ various techniques, such as ultracentrifugation for exosome isolation, next-generation sequencing for analyzing RNA contents, and mass spectrometry for protein profiling. In vitro co-culture systems with host cells are also widely used to assess the functional impact of these exosomes on cellular processes.
Q: What potential applications do Streptococcus pneumoniae exosomes have in biotechnology?
A: These exosomes have potential applications in developing novel vaccine platforms, targeted drug delivery systems, and biomarkers for infection monitoring. Their ability to carry and deliver specific molecules makes them promising tools for therapeutic innovations and diagnostics.
Q: In what ways can studying Streptococcus pneumoniae exosomes advance our understanding of bacterial communication?
A: Investigating these exosomes can reveal how bacteria coordinate activities in response to environmental changes, how they interact with other microbial communities, and how they modulate host immune responses. This knowledge can contribute to the broader field of microbial ecology and pathogenesis.
Q: What are the challenges faced in researching Streptococcus pneumoniae exosomes?
A: Challenges include isolating exosomes in sufficient quantities for analysis, distinguishing them from host-derived exosomes, and understanding the complexity of their cargo and its functional implications. Additionally, replicating in vivo conditions accurately in vitro remains a hurdle.
Q: How do researchers foresee the future of Streptococcus pneumoniae exosome research?
A: Future research may focus on the mechanistic understanding of exosome biogenesis, their specific roles in inter-bacterial communication, and the implications of their RNA and protein cargo in disease progression.
References
-
Mehanny, Mina et al. "Streptococcal Extracellular Membrane Vesicles Are Rapidly Internalized by Immune Cells and Alter Their Cytokine Release." Frontiers in immunology vol. 11 80. Distributed under Open Access license CC BY 4.0. The image was modified by revising the title. https://doi.org/10.3389/fimmu.2020.00080
-
Yerneni, Saigopalakrishna S et al. "Pneumococcal Extracellular Vesicles Modulate Host Immunity." mBio vol. 12,4 (2021): e0165721. Distributed under Open Access license CC BY 4.0.The image was modified by extracting and using only Part C of the original image and revising the title. https://doi.org/10.1128/mbio.01657-21
For Research Use Only. Cannot be used by patients.
Related Services:

 Fig.1 Cellular uptake of Streptococcus pneumoniae-derived exosomes increases with time.1
Fig.1 Cellular uptake of Streptococcus pneumoniae-derived exosomes increases with time.1
 Fig.2 Streptococcus pneumoniae-derived exosomes facilitated host cell recruitment.2
Fig.2 Streptococcus pneumoniae-derived exosomes facilitated host cell recruitment.2