In the past few decades, molecular diagnostics has made great efforts to develop rapid multi analysis methods. New diagnostic techniques have been developed due to advances in the sequencing of the human genome and all encoded proteins. Among various technological solutions for diagnostic applications, array technology has attracted the attention of a wide range of research groups and industries, and greatly affected the methods of disease discovery and genome understanding.
Introduction of Microarray Technique
The microarray consists of a series of ordered probes, which include nucleic acids, nucleic acid analog (such as peptide nucleic acids (PNA), locked nucleic acid (LNA)), proteins, carbohydrates, tissues, cells and polymers. Most microarrays currently in use contain hundreds to thousands of probes. The value of this technology is that it allows highly parallel measurement and has the advantages of high-throughput, miniaturization and high speed. This has led to important applications of microarray technology in the development of medical research, drug resistance, pharmacogenomics and molecular diagnosis.
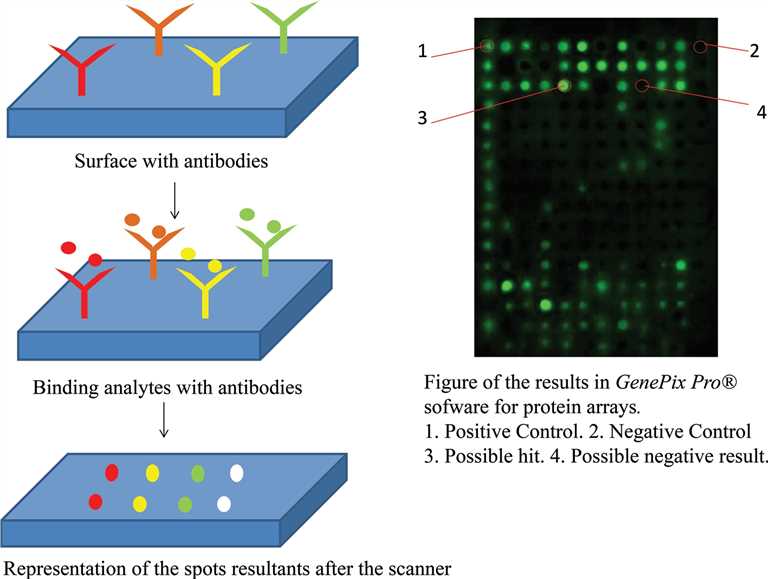 Fig.1 Overall experimental steps in microarray technology.1,3
Fig.1 Overall experimental steps in microarray technology.1,3
Introduction of Array Technique
Among all kinds of in vitro diagnostic (IVD) methods, array technology is the method that provides the highest throughput for the determination of many analytes in relatively few samples, and is listed as one of the most important detection technology modules. Creative Biolabs provides a variety of IVD testing services based on array technology. In summary, there are two main parts:
The liquid array method involves capture probes immobilized on beads, which can be sorted using a flow cytometer. Liquid array diagnosis uses short DNA duplex, in which one oligonucleotide is labeled with a fluorophore and the other is labeled with a quenched molecule in the presence of target DNA. The novelty of this method is to detect multiple fluorescence quenching events in a single tube complex reaction by combining multiple double melting curves and multiple detection channels on qPCR.
The solid array consists of synthetic oligonucleotides or peptides (capture probes) immobilized on a solid substrate (such as a glass slide or nitrocellulose membrane). The number of unique capture probes on a single array can range from 100 on a low-density printed array to >1 million on an in situ-synthesized high-density array. Due to the large number of probes, these arrays are most commonly used for whole-genome expression profiling or other whole-protein group comparisons.
Features
- Quality assurance:
All of our array technology testing services are managed by mature and strict quality assurance measures.
- Expert support:
We have experts in the field of IVD and decades of experience from project design to delivery, providing the most comprehensive testing services for every testing project.
- Customized service:
For different testing requirements, we provide customized high-quality products and services to ensure batch consistency.
If you are interested in our service, please contact us for your exclusive solution.
Published Data
1. Microsphere-Based Suspension Array Assays for Hendra and Nipah Virus Detection and Differentiation
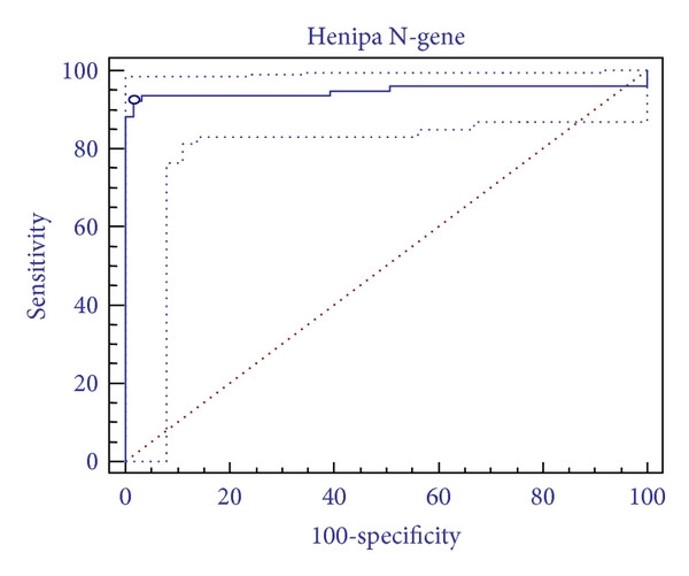 Fig.2 Diagnostic performance characteristics of microsphere array assays.2,3
Fig.2 Diagnostic performance characteristics of microsphere array assays.2,3
Microsphere suspension array systems allow the simultaneous fluorescent detection of multiple nucleotide targets in a single reaction. Researchers used commercially available oligo-tagged microspheres to develop multiplexed assays for detecting and differentiating Hendra virus (HeV) and Nipah virus (NiV), two zoonotic paramyxoviruses transmitted by bats and of increasing concern for human and veterinary medicine. The assays targeted multiple regions within the genes encoding the nucleoprotein (N) and phosphoprotein (P). Specificities and sensitivities were evaluated using reference isolates, samples from experimentally infected horses, and archival veterinary diagnostic submissions, with the results being compared to a validated qPCR method. The microsphere array assays achieved clear differentiation between HeV and NiV, with HeV detection sensitivity comparable to qPCR, demonstrating both high specificity and sensitivity. These assays proved useful for detecting HeV in Australian horses and are intended to be incorporated into higher multiplex assay panels. This will facilitate the development of syndrome-based assay panels for broader disease surveillance in horse, bat, pig, and human populations.
2. Protein Microarray-Based Personalized Proteome Analysis Using Individual Patient Samples
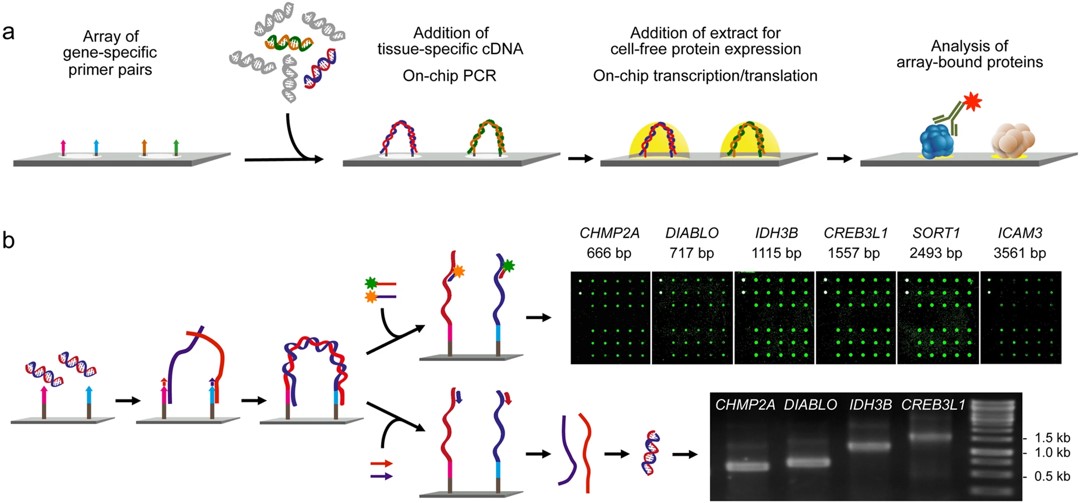 Fig.3 Principle and performance of the process of producing personalized protein microarrays.4
Fig.3 Principle and performance of the process of producing personalized protein microarrays.4
Advances in DNA sequencing have allowed routine analysis of individual patient genomes. To analyze tissue protein content in a similar way, in this study, researchers developed a method for creating microarrays that represent full-length proteins encoded in specific samples, capturing variations such as mutations or splice alterations. Total RNA isolates were used to copy each transcript to a designated array location via an on-chip polymerase elongation reaction, followed by in situ cell-free transcription and translation. These microarrays enable parallel analysis of protein structure and interaction variations that are specific to individual samples, offering insights into sample-specific molecular characteristics.
References
- Juanes-Velasco, Pablo, et al. "Microarrays as platform for multiplex assays in biomarker and drug discovery." Rapid Test-Advances in Design, Format and Diagnostic Applications (2018).
- Foord, Adam J., et al. "Microsphere suspension array assays for detection and differentiation of Hendra and Nipah viruses." BioMed Research International 2013.1 (2013): 289295.
- Distributed under Open Access license CC BY 3.0, without modification.
- Syafrizayanti, et al. "Personalised proteome analysis by means of protein microarrays made from individual patient samples." Scientific reports 7.1 (2017): 39756. Distributed under Open Access license CC BY 4.0, without modification.
For Research Use Only.