Fragment based Screening
Fragment-based screening has proven to be a viable complementary approach to high throughput screening, and it has shown increasing promise in drug discovery. Creative Biolabs has built well-established binding assays for fragment-based hit identification. With sensitive label-free detection and the ability to detect weak interactions for small compounds (less than 250 Da), we are able to provide selective data for hit identification.
The physicochemical properties of fragment libraries are driven by three features. First, the “fragment space” is smaller than the “chemical space”, so it can be more efficiently probed with a relatively small library. Second, the binding efficiency is likely to be higher. Fragment library has low chemical complexity and exhibits high ligand efficiency when comparing larger elaborated drug-like ligands. Third, the hit rates for fragment-based screening appear to be higher, typically 3-10%. It may be useful to use two different screening technologies and then focus on compounds that are active in both assays.
Screening Methodologies
Surface Plasmon Resonance (SPR) Spectrometry
SPR is a powerful tool for studying biomolecular interactions in a sensitive and label-free detection format. It is capable of screening tens of thousands of compounds in a reasonable time frame. This technique only needs small amounts of protein and can provide dissociation constants (Kd values) and, for tighter binders, on-rates and off-rates. However, with any SPR experiment, either protein or ligand must be immobilized. Creative Biolabs currently offers SPR service using either our specially selected fragment library or your own library. We will help you determine suitable immobilization conditions.
Biolayer Interferometry (BLI)
BLI is a methodology that measures changes generated from visible light reflected from an optical layer and a biolayer containing proteins of interest. Like SPR, biolayer interferometry requires the proteins being immobilized on a solid support.
Nuclear Magnetic Resonance (NMR) Spectroscopy
NMR has become a preferred method for identifying fragments that specifically bind to a protein or nucleic acid targets for even weak ligands. The method can detect nM to mM interactions and provides structural information of the binding site. However, NMR requires a relatively large amount of protein and has size/solubility limitations for proteins (solubility between 0.1-1 mM).
Isothermal Titration Calorimetry (ITC)
ITC is a physical technique used to determine the heat changes caused by interactions in solutions. It is typically used as a secondary screening method. ITC gives not only the binding affinity but also the thermodynamics of the binding.
X-Ray Crystallography
The advantage of this approach is that compounds can be seen and evaluated. Therefore, it has low false positives. In addition, it may be possible to enhance binding affinity by using modeling techniques or purchasing related analogues. However, X-ray crystallography is a very time and resource consuming project in which a large protein (10-50 mg of the target protein with a purity of over 95%) amount is needed. Also, fragment ligands need soluble in the crystallization medium.
Thermal Shift Assay
A thermal shift assay, also known as differential scanning calorimetry, refers to quantifying the variation in thermal denaturation temperature of a protein in different surroundings. The rank order of ligand binding and the estimate of the binding energy can be obtained by comparing the Tm values of the apoprotein and protein-ligand complexes. This can be measured by binding to a fluorescent marker.
Mass Spectrometry (MS)
MS has been recognized as a rapid, sensitive, and high throughput method to directly investigate protein-fragment interactions. It only requires a tiny amount of proteins and does not need immobilization or conjugation. However, it may lead to false positives due to aggregation as it requires the absence of detergent.
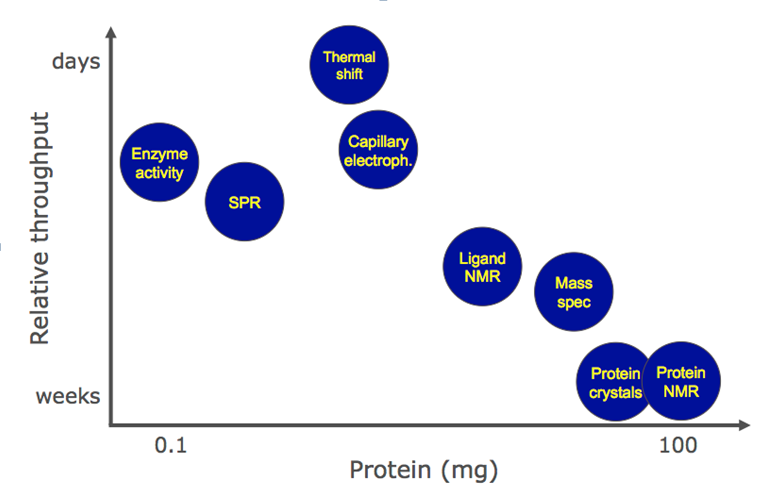 Fig. 1 Comparison of different fragment-based screening methods.1
Fig. 1 Comparison of different fragment-based screening methods.1
For more detailed information, please feel free to contact us or directly sent us an inquiry.
Reference
- Cambridge MedChem Consulting 2012, Fragment-Based Screening, digital image, accessed 20 September 2016, {http://www.cambridgemedchemconsulting.com/resources/hit_identification/fragment_based_screening.html}. Distributed under Open Access license CC BY 3.0, without modification.
For Research Use Only.
